Volume 7, Number 4—August 2001
THEME ISSUE
West Nile Virus
West Nile Virus
Rapid Determination of HLA B*07 Ligands from the West Nile Virus NY99 Genome
Figure 1
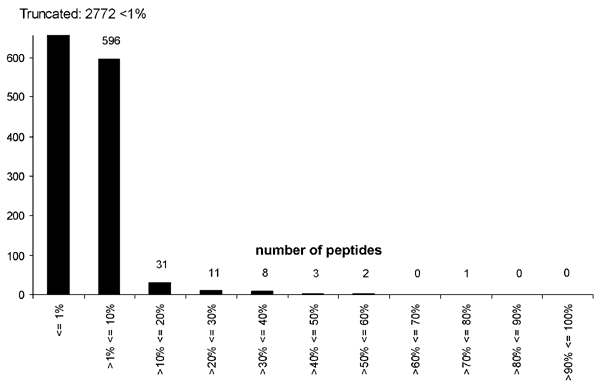
Figure 1. . . Distribution of scores for the complete set of 3,424 peptides obtained by parsing the West Nile (WN) virus genome into 10 amino-acid long peptides, each overlapping by 9 amino acids, as scored on the EpiMatrix motif for HLA B*07. Peptides with estimated binding potential (EBP) scores >7 and <50 with the HLA B*07 motif are highly likely to bind to HLA B*07 in T2 B7 assays and to stimulate T cells. WN virus peptides with EBP scores between 20 and 50 were considered for study.
Page created: April 27, 2012
Page updated: April 27, 2012
Page reviewed: April 27, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.