Volume 8, Number 7—July 2002
Research
First Human Isolate of Hantavirus (Andes virus) in the Americas
Figure 2
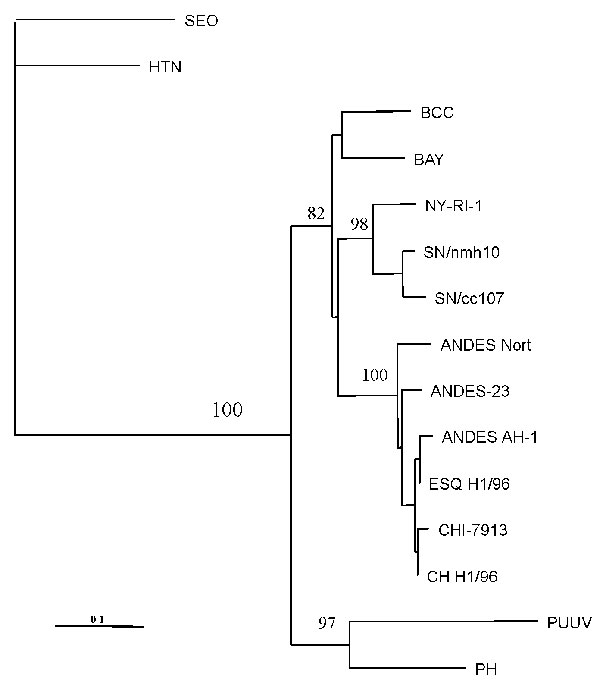
Figure 2. Maximum-parsimony tree analysis comparing S nucleotide sequence of CHI-7913 virus with homologous sequences of previously characterized hantaviruses. Nucleotide sequences examined correspond to positions 22–359 of antigenome-sense sequence of nucleoprotein (N) gene. Sequences were analyzed by the maximun parsimony method with the Clustal W and PHYLIP packages (15). The minimal length trees shown were supported as the majority rule consensus tree in 500 replicates. The bootstrap replicates supporting each node are indicated. References and GenBank accession numbers for the sequences used in S segment comparisons are BCC (16) L39949; BAY (17) L36929; NY strain RI-1 (18), U09488; SN strain cc107 (19), L33683; SN strain nmh10 (20), L25784; PH strain PH-1 (21), and M34011; Puumala strains Sotkamo (22), X61035; Seoul (SEO) strain sr-11 (23), and M34881; Hantaan (HTN) strain 76-118 (24), M14626; Andes strain AH-1 (14), AF004660; ESQ H-1/96 (14), AF005948; CH H-1/96 (14), AF 005947; AND Nort (strain unpublished) AF325966; and Andes strain 23 (AF291702).
References
- Schmaljohn C, Hjelle B. Hantavirus: a global disease problem. Emerg Infect Dis. 1997;3:95–104.PubMedGoogle Scholar
- Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Felmann H, Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1994;262:832–6.
- Duchin JS, Koster F, Peters CJ, Simpson GL, Tempest B, Zaki SR, Hantavirus pulmonary syndrome: a clinical description of 17 patients with a newly recognized disease. N Engl J Med. 1994;330:949–55. DOIPubMedGoogle Scholar
- Hallin G, Simpson S, Crowell R, James DS, Koster FT, Mertz GJ, Cardiopulmonary manifestations of hantavirus pulmonary syndrome. Crit Care Med. 1996;24:252–8. DOIPubMedGoogle Scholar
- Chilean Ministry of Health. Epidemiologic report of Hantavirus in Chile. Santiago, Chile: the Ministry; 2001.
- Padula PJ, Edelstein A, Miguel SD, Lopez NM, Rossi CM, Rabinovich RD. Hantavirus pulmonary syndrome outbreak in Argentina: molecular evidence for person-to-person transmission of Andes virus. Virology. 1998;241:323–30. DOIPubMedGoogle Scholar
- Terajima M, Hendershot JD, Kariwa H, Koster FT, Hjelle B, Goade D, High levels of viremia in patients with the hantavirus pulmonary syndrome. J Infect Dis. 1999;180:2030–4. DOIPubMedGoogle Scholar
- Mertz GJ, Hjelle BL, Williams TM, Koster FT. Host responses in the Hantavirus cardiopulmonary syndrome. In: Saluzzo JF, Dodet B, editors. Factors in the emergence and control of rodent-borne viral diseases. Paris: Elsevier; 1999. p. 133–7.
- Bharadwaj M, Nofchissy R, Goade D, Koster F, Hjelle B. Humoral immune responses in the hantavirus cardiopulmonary syndrome. J Infect Dis. 2000;182:43–8. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention and National Institutes of Health. Biosafety in microbiological and biomedical laboratory (BMBL). 4th edition. Washington: U.S. Government Printing Office; 1999.
- Rossi C, Ksiazek T. Virus detection and identification with serological tests.2. Enzyme-linked immunosorbent assay (ELISA). In: Lee H W, Calisher C, Schmaljohn C, editors. Manual of hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome. Seoul: WHO Collaborating Centre for Virus Reference and Research (Hantavirus); 1999. p. 87–91.
- Lee HW. Virus isolation. In: Lee HW, Calisher C, Schmaljohn C, editors. Manual of hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome. Seoul: WHO Collaborating Centre for Virus Reference and Research (Hantavirus); 1999. p. 74–9.
- Gallo D, Penning LM, Hanson CV. Detection and differentiation of antibodies to human T-cell lymphotropic virus types I and II by immunofluorescence method. J Clin Microbiol. 1991;29:2345–7.PubMedGoogle Scholar
- Lopez N, Padula P, Rossi C, Miguel S, Edelstein A, Ramirez E, Genetic characterization and phylogeny of Andes virus and variants from Argentina and Chile. Virus Res. 1997;50:77–84. DOIPubMedGoogle Scholar
- Kuhner MK, Felsenstein J. A simulation comparison of phylogeny algorithms under equal and unequal evolutionary rates. Mol Biol Evol. 1994;11:459–68.PubMedGoogle Scholar
- Ravkov EV, Rollin PE, Ksiazek TG, Peters CJ, Nichol ST. Genetic and serologic analysis of Black Creek Canal virus and its association with human disease and Sigmodon hispidus infection. Virology. 1995;210:482–9. DOIPubMedGoogle Scholar
- Morsunov S, Feldmann H, Spiropoulou CF, Semenova VA, Rollin PE, Ksiazek TG, A newly recognized virus associated with a fatal case of hantavirus pulmonary syndrome in Louisiana. J Virol. 1995;69:1980–3.PubMedGoogle Scholar
- Hjelle B, Krolikowski J, Torrez-Martinez N, Chavez-Giles F, Vanner C, Laposata E. Phylogenetically distinct hantavirus implicated in a case of hantavirus pulmonary syndrome in the northeastern United States. J Med Virol. 1995;46:21–7. DOIPubMedGoogle Scholar
- Schmaljohn AL, Li D, Negley DL, Bressler DS, Turrell MJ, Korch GW, Isolation and initial characterization of a newfound hantavirus from California. Virology. 1995;206:963–72. DOIPubMedGoogle Scholar
- Spriropoulou CF, Morzunov S, Feldmann H, Sanchez A, Peters CJ, Nichol ST. Genome structure and variability of a virus causing hantavirus pulmonary syndrome. Virology. 1994;200:715–23. DOIPubMedGoogle Scholar
- Parrington MA, Kang YC. Nucleotide sequence analysis of the S genomic segment of Prospect Hill virus: comparison with the prototype hantavirus. Virology. 1990;175:167–75. DOIPubMedGoogle Scholar
- Vapalahti O, Kallio-Kokko H, Salonen EM, Brummer-Korvenkontio M, Vaheri A. Cloning and sequencing of Puumala virus Sotkamo strain S and M RNA segments: evidence for strain variation in hantavirus and expression of the nucleocapsid protein. J Gen Virol. 1992;73:829–38. DOIPubMedGoogle Scholar
- Arikawa J, Lapenotiere HF, Iacono-Connors L, Wang M, Schmaljohn CS. Coding properties of the S and M genome segments of Sapporo rat virus: comparison to other causative agents of hemorrhagic fever with renal syndrome. Virology. 1990;176:114–25. DOIPubMedGoogle Scholar
- Schmaljohn CS, Jennings AL, Hay J, Dalrymple JM. Coding strategy of the S genome segment of Hantaan virus. Virology. 1986;155:633–43. DOIPubMedGoogle Scholar
- Xiao SY, Leduc JW, Chu YK, Schmaljohn CS. Phylogenetic analyses of virus isolates in the genus Hantavirus, family Bunyaviridae. Virology. 1994;198:205–17. DOIPubMedGoogle Scholar
- Juto P, Elgh F, Ahlm C, Alexeyev OA, Edlund K, Lundkvist A, The first human isolate of Puumala virus in Scandinavia as cultured from phytohemagglutinin stimulated leucocytes. J Med Virol. 1997;53:150–6. DOIPubMedGoogle Scholar
- Gu XS, Song ZB, Jin ZW, Meng GR, Zhang CA, Yan DY, Isolation of a strain of Hantaan virus from peritoneal exudate cells of a patient with hemorrhagic fever with renal syndrome. Chin Med J. 1990;103:455–9.PubMedGoogle Scholar
- Hjelle B, Spiropoulou CF, Torrez-Martinez N, Morzunov S, Peters CJ, Nichol ST. Detection of Muerto Canyon virus RNA in peripheral blood mononuclear cells from patients with hantavirus pulmonary syndrome. J Infect Dis. 1994;170:1013–7.PubMedGoogle Scholar