Volume 9, Number 1—January 2003
Dispatch
Enteropathogenic Escherichia coli O157 Strains from Brazil
Cite This Article
Citation for Media
Abstract
We describe two serogroup O157 Escherichia coli strains from Brazilian infants with diarrhea. A variety of assays indicate that these strains belong to the enteropathogenic, not the enterohemorrhagic, pathotype. These strains possess a novel bfpA allele encoding the type IV pilin characteristic of typical enteropathogenic E. coli strains. Our results emphasize the pitfalls of classifying pathogenic E. coli by serogroup.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is the serotype most commonly associated with hemorrhagic colitis and the hemolytic uremic syndrome (1). EHEC strains share with enteropathogenic E. coli (EPEC), a leading cause of infant diarrhea in developing countries, the ability to induce the attaching and effacing effect on host cells. This property is specified by a pathogenicity island that includes the eae gene encoding the outer membrane adhesin intimin. EPEC are defined by this attaching and effacing phenotype or, at a molecular level, by the presence of the eae gene and the absence of the genes for Shiga toxins (2). Typical EPEC strains have a large plasmid that encodes bundle-forming pili and the localized adherence phenotype, while atypical strains lack these properties. EHEC differ from EPEC in that they produce Shiga toxins but not bundle-forming pili.
Here we describe two strains of the O157 serogroup identified as part of an ongoing epidemiologic survey of pathogenic E. coli in Brazil (3). Strain SC373/2 was isolated from a 9-month-old infant in Joinville, Santa Catarina, in 1997, and RN587/1, from a 7-month-old infant in Natal, Rio Grande do Norte, in 1998. Both patients had acute diarrhea of 7 days’ duration, accompanied by vomiting, fever, and moderate dehydration. These strains were the only enteropathogenic bacteria isolated from the patients’ stools. Rotavirus and cryptosporidia were not detected.
We used a combination of phenotypic assays and DNA sequencing to further characterize these isolates. DNA probe testing was performed by established methods (4). A fragment of the eae gene was amplified by polymerase chain reaction (PCR) and sequenced as described (5). Serotyping was performed at the E. coli Reference Laboratory (University Park, PA). Tests of localized adherence, autoaggregation, and attaching and effacing were performed as described (6,7). The bfpA gene was amplified by PCR and sequenced as described (8). Multilocus sequence typing was performed as described (9).
We found a number of indications that these O157 strains are not EHEC but unexpectedly fall within the EPEC pathotype. First, both strains tested probe negative for the Shiga toxin (stx) genes characteristic of EHEC. Second, both strains carried an eae (intimin) gene, identified by sequencing as an α allele characteristic of the group of strains known as EPEC 1, rather than the γ allele characteristic of O157:H7 EHEC (C.L. Tarr and T.S. Whittam, unpub. data). As expected of both EPEC and EHEC, both strains are capable of attaching and effacing as demonstrated by using the fluorescence actin staining test. E. coli strains that are eae+ and stx– are by definition, EPEC (2). Third, both strains lacked the H7 flagellar antigen, although they were motile. The H antigens were difficult to type; RN587/1 reacts weakly with H8 antiserum. Fourth, both strains rapidly fermented sorbitol, a phenotype lacking in most O157:H7 strains. Fifth, the nucleotide sequences of part of the coding regions of 14 chromosomal loci encoding proteins with housekeeping functions indicated a close relationship of these strains with the EPEC 1 clonal group. The DNA sequences of the two O157 strains were identical over a total of 2,240 codons. Comparison of the multilocus sequence data to the reference strains K-12, EDL-933 (O157:H7; EHEC 1), and E2348/69 (O127:H6; EPEC 1) indicated that the EPEC O157 isolates were most closely related to E2348/69. The percentage of nucleotide difference in the total of 6,720 bp was 0.72 ± 0.10 (E2348/69), 2.66 ± 0.20 (K-12), and 2.95 ± 0.21 (EDL-933). Phylogenetic analysis with these sequences from a diverse collection of strains, including EPEC, EHEC, and other pathotypes, likewise shows that RN587/1 and SC373/2 are much more closely related to EPEC 1 than to EHEC O157 strains (Figure 1). Both strains also tested probe positive for the EPEC adherence factor plasmid, which is associated with localized adherence.
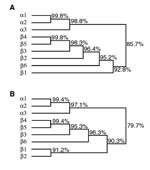
Finally, both strains carried the bfpA gene encoding bundlin, the structural subunit of the bundle-forming pilus expressed by typical EPEC strains but not by EHEC. Both strains are also capable of autoaggregation and localized adherence, the two phenotypes associated with expression of bundle-forming pili. The bfpA gene isolated from these strains has a unique sequence (GenBank accession number AF474407). We call this version of bfpA the β6 allele. When compared to the α and β bfpA alleles described previously (8), the β6 bfpA allele maps between the β1 allele and a cluster of the β2–β5 alleles (Figure 2). The predicted β6 bundlin amino acid sequence maps between the β1 and β2 and the β3–5 pilin proteins.
Our findings emphasize the fact that E. coli with the O157 O antigen are not always EHEC but may belong to other pathotypes. Scotland et al. described Shiga toxin–negative O157:H8 E. coli strains isolated from children with diarrhea (10). These strains were positive for attaching and effacing and localized adherence phenotypes and are therefore EPEC, although, unlike the strains we describe, they were negative for the adherence factor probe. EPEC strains of the O157:H45 serotype have been noted as agents of diarrhea in a large outbreak in Japan and isolated cases in Germany, Japan, and Thailand (11–13). These previously described O157 EPEC strains were not tested for bundlin alleles, and their phylogenetic relationships to other pathogenic E. coli have not been reported. However, the results of several studies have indicated that O157 strains lacking the H7 flagellar antigen and stx genes, which were not necessarily EPEC, are clonally distinct from O157:H7 strains and often from each other (14–16). Such findings suggest that recombination events causing the O antigen gene cluster to specify the O157 antigen have occurred in multiple E. coli lineages, including an EPEC 1 strain of an unknown serotype that is the progenitor of the strains described here. The results described in this study also highlight the pitfalls of classifying pathogenic E. coli by serogroup.
Dr. Blank is a microbiologist with GEO-CENTERS, Inc. and conducts research in the laboratory of Arthur Friedlander at the U.S. Army Medical Research Institute of Infectious Diseases in Frederick, Maryland. Dr. Blank’s research focuses on genetic variations that attenuate the virulence of bacterial pathogens.
Acknowledgments
We are grateful to Wensheng Luo and Xiaolin Wang for technical assistance.
This work was supported by Public Health Service grant (R01 AI-37606) to M.S.D., a National Research Service Award postdoctoral training fellowship (F32 AI-10191) to T.E.B., and in part by the National Institutes of Health through the Enteric Pathogens Research Unit, University of Maryland Medical School (T.S.W.).
References
- Donnenberg MS, Whittam TS. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J Clin Invest. 2001;107:539–48. DOIPubMedGoogle Scholar
- Kaper JB. Defining EPEC. Revista de Microbiologia, Sao Paulo. 1996;27(Suppl1):130–3.
- Scaletsky ICA, Fabbricotti SH, Aranda KR, Morais MB, Fagundes-Neto U. Comparison of DNA hybridization and PCR assays for detection of putative pathogenic enteroadherent Escherichia coli. J Clin Microbiol. 2002;40:1254–8. DOIPubMedGoogle Scholar
- Scotland SM, Smith HR, Cheasty T, Said B, Willshaw GA, Stokes N, Use of gene probes and adhesion tests to characterise Escherichia coli belonging to enteropathogenic serogroups isolated in the United Kingdom. J Med Microbiol. 1996;44:438–43. DOIPubMedGoogle Scholar
- McGraw EA, Li J, Selander RK, Whittam TS. Molecular evolution and mosaic structure of α, β, and γ intimins of pathogenic Escherichia coli. Mol Biol Evol. 1999;16:12–22.PubMedGoogle Scholar
- Donnenberg MS, Nataro JP. Methods for studying adhesion of diarrheagenic Escherichia coli. Methods Enzymol. 1995;253:324–36. DOIPubMedGoogle Scholar
- Anantha RP, Stone KD, Donnenberg MS. Effects of bfp mutations on biogenesis of functional enteropathogenic Escherichia coli type IV pili. J Bacteriol. 2000;182:2498–506. DOIPubMedGoogle Scholar
- Blank TE, Zhong H, Bell AL, Whittam TS, Donnenberg MS. Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia coli strains. Infect Immun. 2000;68:7028–38. DOIPubMedGoogle Scholar
- Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–7. DOIPubMedGoogle Scholar
- Scotland SM, Willshaw GA, Cheasty T, Rowe B. Strains of Escherichia coli O157:H8 from human diarrhoea belong to attaching and effacing class of E. coli. J Clin Pathol. 1992;45:1075–8. DOIPubMedGoogle Scholar
- Schmidt H, Rüssmann H, Karch H. Virulence determinants in nontoxinogenic Escherichia coli O157 strains that cause infantile diarrhea. Infect Immun. 1993;61:4894–8.PubMedGoogle Scholar
- Makino S, Asakura H, Shirahata T, Ikeda T, Takeshi K, Arai K, Molecular epidemiological study of a mass outbreak caused by enteropathogenic Escherichia coli O157:H45. Microbiol Immunol. 1999;43:381–4.PubMedGoogle Scholar
- Echeverria P, Orskov F, Orskov I, Knutton S, Scheutz F, Brown JE, Attaching and effacing enteropathogenic Escherichia coli as a cause of infantile diarrhea in Bangkok. J Infect Dis. 1991;164:550–4.PubMedGoogle Scholar
- Whittam TS, Wilson RA. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect Immun. 1988;56:2467–73.PubMedGoogle Scholar
- Feng P, Lampel KA, Karch H, Whittam TS. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–3. DOIPubMedGoogle Scholar
- Tarr PI, Schoening LM, Yea YL, Ward TR, Jelacic S, Whittam TS. Acquisition of the rfb-gnd cluster in evolution of Escherichia coli O55 and O157. J Bacteriol. 2000;182:6183–91. DOIPubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 9, Number 1—January 2003
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Michael S. Donnenberg, Division of Infectious Diseases, Department of Medicine, University of Maryland School of Medicine, Baltimore, Maryland, 21201, USA; fax: 410-706-8700
Top