Volume 11, Number 1—January 2005
Dispatch
A Novel Paramyxovirus?
Figure 1
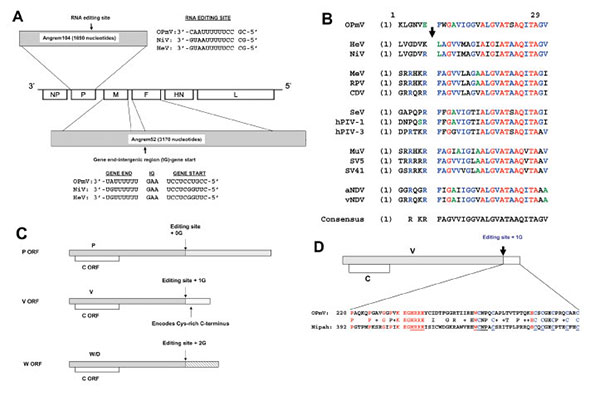
Figure 1. Angrem52 and Angrem104 appear to be paramyxovirus genes. A) Gene positions of a generic paramyxovirus and predicted genome position of Angrem104 (top), the phosphoprotein (P) gene, Angrem52 (bottom), the matrix protein (M) and fusion protein (F) genes. A potential editing site (nucleotides 783–795), which might allow production from the OPmV P gene of V and W/D proteins, is shown in genomic (negative) sense aligned with the proposed editing sites of Nipah virus (NC_002728) (1) and Hendra viruses (NC_001906). The full-length P open reading frame (ORF) was obtained by inserting an additional nucleotide in the reported Angrem104 sequence (see text). Angrem52 is predicted to be a “read-through” product of the M and F genes, a novel paramyxovirus. The full-length F ORF was obtained by making 5 changes to the reported Angrem52 sequence (see text). Putative gene-end, intergenic (IG), and gene-start transcription regulatory signals lying between OPmV M and F genes are shown aligned to the corresponding signals from Nipah and Hendra virus (shown in genomic sense [12]). B) The putative OPmV F protein contains a fusion peptide. The sequences surrounding the F protein cleavage sites, including most fusion peptides, of several paramyxoviruses, including putative OPmV, were aligned by using the AlignX program of the Vector NTI6 software package. The arrow indicates the cleavage site. Residues in red are absolutely conserved. Residues in blue are conserved in most sequences. C) Putative genome organization of the putative OPmV P gene, allowing translation of P, V, W, and C ORFs. D) Alignment of cysteine-rich carboxy-termini of the putative OPmV and Nipah virus V proteins. The conserved carboxy-terminal regions of the V proteins were aligned by using the AlignX program of the Vector NTI6 software package. Conserved residues are indicated in red, except for conserved cysteines, which are in blue. Underlined residues are conserved among all paramyxovirus V proteins.
References
- Gallo RC, Montagnier L. The discovery of HIV as the cause of AIDS. N Engl J Med. 2003;349:2283–5. DOIPubMedGoogle Scholar
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–62. DOIPubMedGoogle Scholar
- Jia XY, Briese T, Jordan I, Rambaut A, Chi HC, Mackenzie JS, Genetic analysis of West Nile New York 1999 encephalitis virus. Lancet. 1999;354:1971–2. DOIPubMedGoogle Scholar
- Peiris JS, Yuen KY, Osterhaus AD, Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–41. DOIPubMedGoogle Scholar
- Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH, A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004;101:6212–6. DOIPubMedGoogle Scholar
- Klenk H-D, Slenczka W, Feldmann H. Marburg and Ebola viruses. In: Webster RG, Granoff A, editors. Encyclopedia of virology. Vol. 2. New York: Academic Press; 1994. p. 827–31.
- Field H, Young P, Yob JM, Mills J, Hall L, Mackenzie J. The natural history of Hendra and Nipah viruses. Microbes Infect. 2001;3:307–14. DOIPubMedGoogle Scholar
- Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–5. DOIPubMedGoogle Scholar
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–24. DOIPubMedGoogle Scholar
- Liang X, Zhang H, Zhou A, Hou P, Wang H. Screening and identification of the up-regulated genes in human mesangial cells exposed to angiotensin II. Hypertens Res. 2003;26:225–35. DOIPubMedGoogle Scholar
- Liang X, Zhang H, Zhou A, Wang H. AngRem104, an angiotensin II-induced novel upregulated gene in human mesangial cells, is potentially involved in the regulation of fibronectin expression. J Am Soc Nephrol. 2003;14:1443–51. DOIPubMedGoogle Scholar
- Harcourt BH, Tamin A, Ksiazek TG, Rollin PE, Anderson LJ, Bellini WJ, Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology. 2000;271:334–49. DOIPubMedGoogle Scholar
- Lamb RA, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Field’s virology. Vol. 1, 4th ed. Philadelphia: Lippincott Williams and Wilkins; 2001. p. 1305–40.
- Randall RE, Russell WC. Paramyxovirus persistence: consequences for host and virus. In: Kingsbury DW, editor. The paramyxoviruses. New York: Plenum Press; 1991.
- Klenerman P, Hengartner H, Zinkernagel RM. A non-retroviral RNA virus persists in DNA form. Nature. 1997;390:298–301. DOIPubMedGoogle Scholar