Volume 13, Number 12—December 2007
Research
Epidemiology and Molecular Virus Characterization of Reemerging Rabies, South Africa
Figure 4
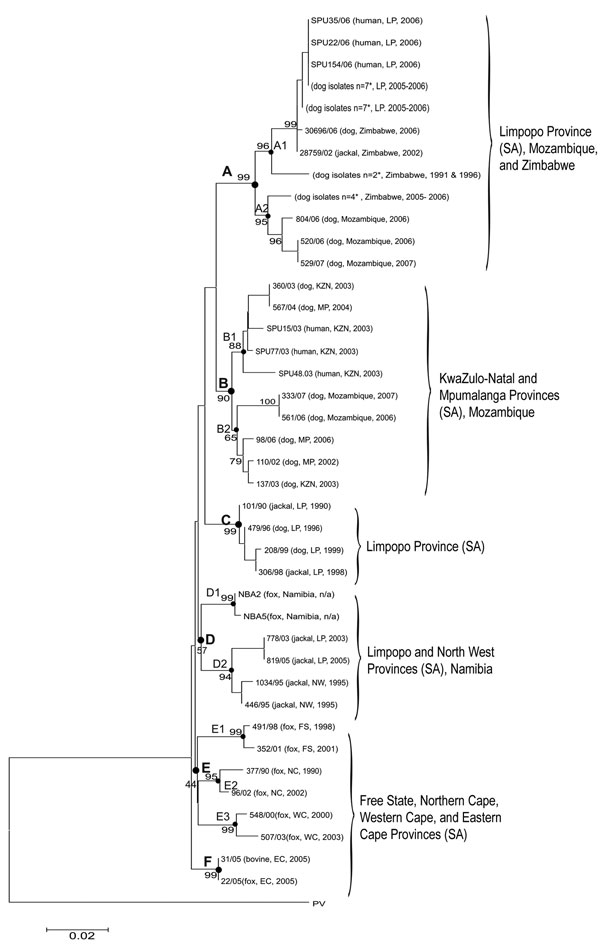
Figure 4. Neighbor-joining tree of canid rabies viruses from humans and animals from Limpopo (LP), Mpumalanga (MP), North West (NW), Free State (FS), Eastern Cape (EC), Northern Cape (NC), KwaZulu-Natal (KZN), and Western Cape (WC) Provinces of South Africa (SA) and neighboring countries of Zimbabwe, Mozambique, and Namibia. The Pasteur virus strain (PV) was used as the reference strain in the sequence alignment. Horizontal scales represent the evolutionary distance; vertical lines are for clarification purposes only. The scale bar indicates nucleotide substitutions per site. Viruses are identified by a laboratory reference number, source animal, locality of origin, and year of isolation. A–F represent virus lineages supported by bootstrap values of >70%; sublineages are indicated numerically. *Identical strains.