Volume 13, Number 9—September 2007
Dispatch
Coronavirus Antibodies in African Bat Species
Figure 1
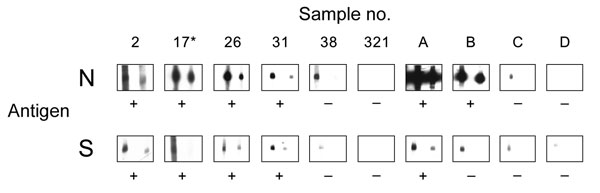
Figure 1. Results of Western blot analysis with recombinant severe acute respiratory syndrome–associated coronavirus (SARS-CoV) nucleocapsid (N) and spike (S) protein. Shown are examples for SARS-CoV ELISA–positive (2, 17, 26, 31) and –negative (38, 321) bat serum specimens tested using full-length recombinant SARS-CoV N and a fragment of the S protein (amino acids 318–510). Serum specimens were diluted 1:2,500 (left strips) and 1:5,000 (right strips). Secondary detection was performed by incubating the nitrocellulose strips with horseradish peroxidase (HRP)–labeled goat-antibat immunoglobulin (Ig) (Bethyl, Montgomery, AL, USA) (1:10,000). For chemiluminescence, SuperSignal Dura substrate (Pierce, Rockford, IL, USA) was added and films exposed for 1 min. Serum 17* was used as a reference for comparing blots. For evaluation purposes, strips were also incubated with human SARS-CoV–positive (A, B) and –negative serum specimens C and D (HCoV-NL63 positive) at the same dilutions, using goat-antihuman Ig HRP (1:20,000) for secondary detection. Serum specimens that produced signals at a dilution of 1:5,000 were recorded as positive (+).