Volume 14, Number 4—April 2008
Research
Detection and Prevalence Patterns of Group I Coronaviruses in Bats, Northern Germany
Figure 2
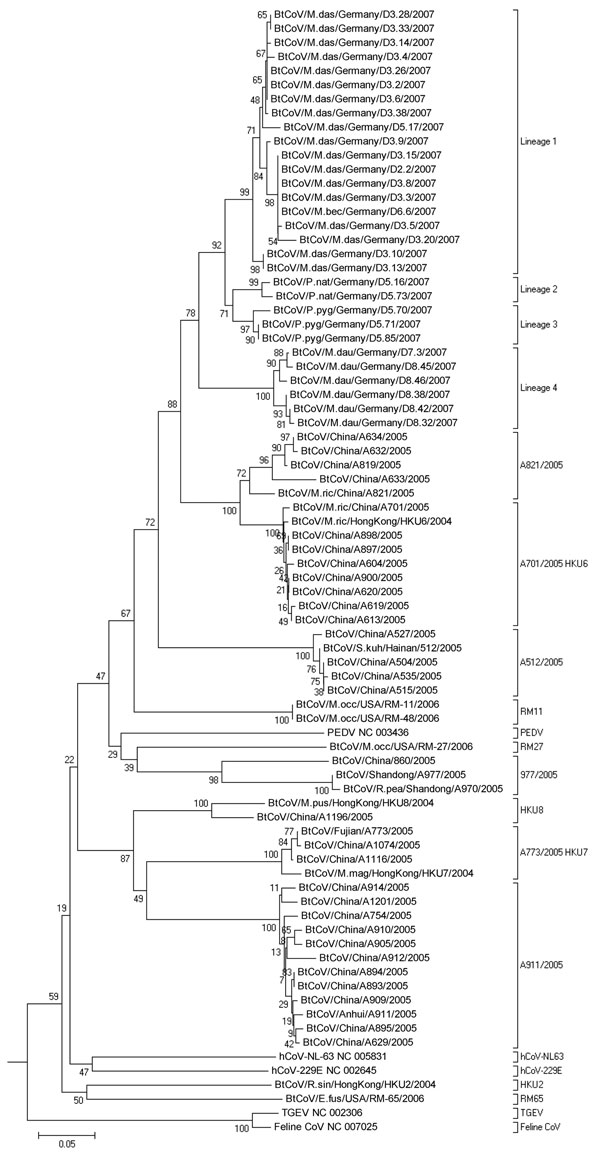
Figure 2. Phlyogenetic analysis of northern German bat coronaviruses (CoV) (lineages 1–4) and related group I CoVs from bats and other mammals. Analyses were conducted in MEGA4 (32), by using the neighbor-joining algorithm with Kimura correction and a bootstrap test of phylogeny. Numbers at nodes denote bootstrap values as percentage of 1,000 repetitive analyses. The phylogeny is rooted with a Leopard CoV, ALC/GX/F230/06 (33). The column on the right shows bat CoV prototype strain names or the designations of type strains of established mammalian CoV species.
Page created: July 16, 2010
Page updated: July 16, 2010
Page reviewed: July 16, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.