Volume 14, Number 9—September 2008
Letter
West Nile Virus in Golden Eagles, Spain, 2007
Figure
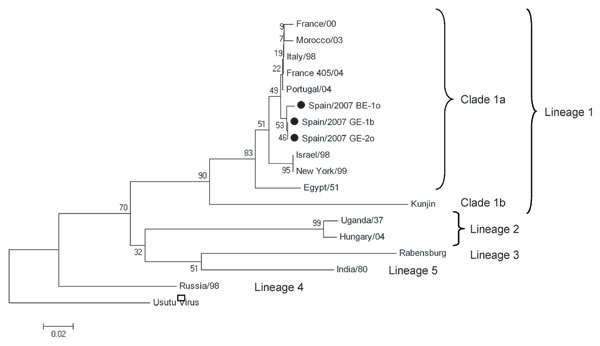
Figure. Phylogenetic tree of 18 partial nonstructural protein 5 West Nile virus nucleotide sequences (171 nt for each isolate, except 126 nt available for the Portugal/04 isolate) constructed with MEGA version 4 software (www.megasoftware.net). The optimal tree was inferred by using the neighbor-joining method. The percentage of successful bootstrap replicates (N = 1,000) is indicated at nodes. Evolutionary distances were computed with the Kimura 2-parameter method (with gamma correction). All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons. Branch lengths are proportional to the number of nucleotide changes (genetic distances). Scale bar shows number of base substitutions per site. Isolates sequenced in this study are indicated by solid circles. GenBank accession nos. are as follows: France/00 (AY268132), Morocco/03 (AY701413), Italy/98 (AF404757), France 405/04 (DQ786572), Portugal/04 (AJ965630), Israel/98 (AF481864), New York/99 (DQ211652), Egypt/51 (AF260968), Kunjin (D00246), Uganda/37 (M12294), Hungary/04 (DQ116961), Rabensburg (AY765264), India/80 (DQ256376), Russia/98 (AY277251), and Usutu virus (NC_006551) (outgroup).