Volume 15, Number 4—April 2009
Dispatch
Concurrent Chikungunya and Dengue Virus Infections during Simultaneous Outbreaks, Gabon, 2007
Figure 2
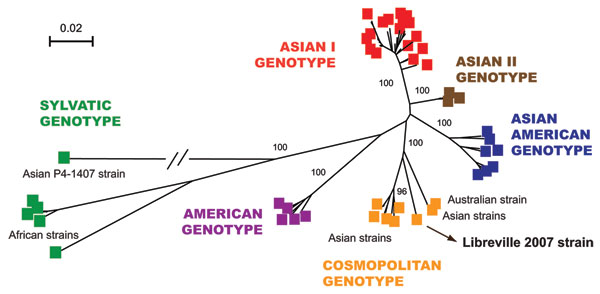
Figure 2. Phylogenetic relationships among dengue-2 virus (DENV-2) isolates based on full-length sequences (10,695 nt). A total of 85 DENV-2 genomes were compared with the human isolate obtained during the Gabon outbreak. Phylogeny was inferred by using neighbor-joining analysis. A neighbor-joining tree was constructed by using MEGA version 3.2 (www.megasoftware.net) with the Kimura 2-parameter corrections of multiple substitutions. Reliability of nodes was assessed by bootstrap resampling with 1,000 replicates. Branches are scaled according the number of substitutions per site, and the branch leading to the Thailand 94 strain was shortened for convenience. Bootstrap values are shown for major key nodes.
Page created: December 10, 2010
Page updated: December 10, 2010
Page reviewed: December 10, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.