Volume 15, Number 5—May 2009
Dispatch
Genotype Distribution and Sequence Variation of Hepatitis E Virus, Hong Kong
Figure
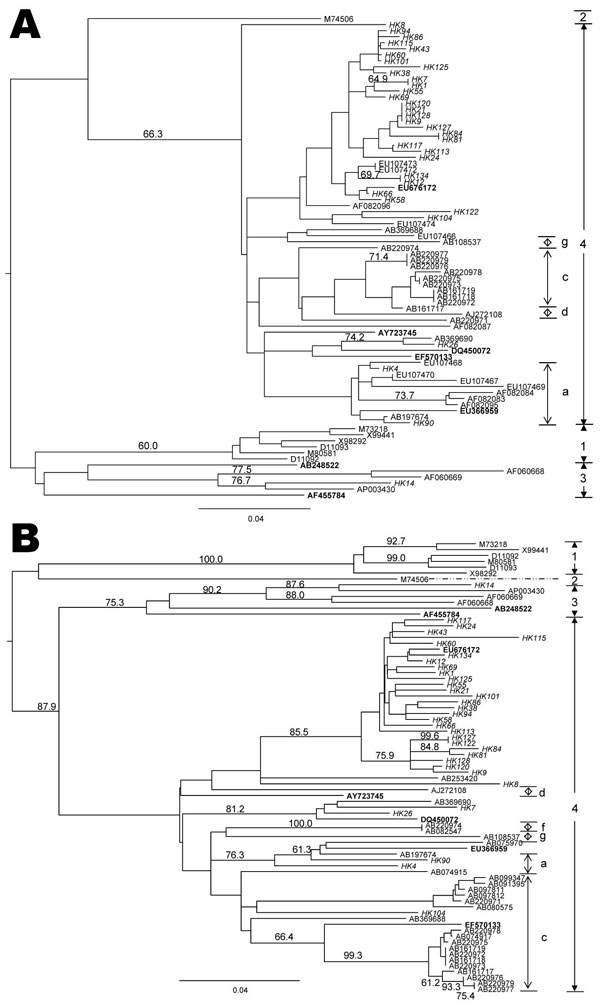
Figure. Phylogenetic tree showing the relationship of hepatitis E virus (HEV) isolates from Hong Kong. Trees were constructed by the neighbor-joining method based on the partial nucleotide sequence of the open reading frame (ORF) 2 (A) and ORF1 (B) regions of HEV samples. Genotypes are indicated by numbers and subtypes by letters on the right. Branch lengths are proportional to genetic distance. Scale bars indicate 0.04 nt substitutions per position. Bootstrap values for the various branches are shown as percentages of trees obtained from 1,000 resamplings of the data. Sequences used for phylogenetic analysis were isolates from genotype 1: Burma (GenBank accession nos. M73218; D10330), Pakistan (M80581), India (X98292; X99441), and China (D11092; D11093); genotype 2: Mexico (M74506); genotype 3: USA (AF060668; AF060669), Japan (AP003430); and genotype 4: China (AB108537; AB197674; AF082083; AF082084; AF082087; AF082095; AF082096; AJ272108; EU107466-74), Japan (AB074915; AB074917; AB080547; AB091395; AB097812; AB099347; AB161717-19; AB220971-79; AB253420; AB369688; AB369690). Branches of swine HEV genotype 3 sequences (AB238522; AF455784 [experimentally infected swine]) and swine HEV genotype 4 sequences (AY723745; DQ450072; EF570133; EU366959; EU676172) are included in the analysis. Accession numbers in boldface are swine isolates. All isolates from the current study have a prefix HK followed by a number in italics.