Volume 15, Number 9—September 2009
Dispatch
Genetic Differences between Avian and Human Isolates of Candida dubliniensis
Figure
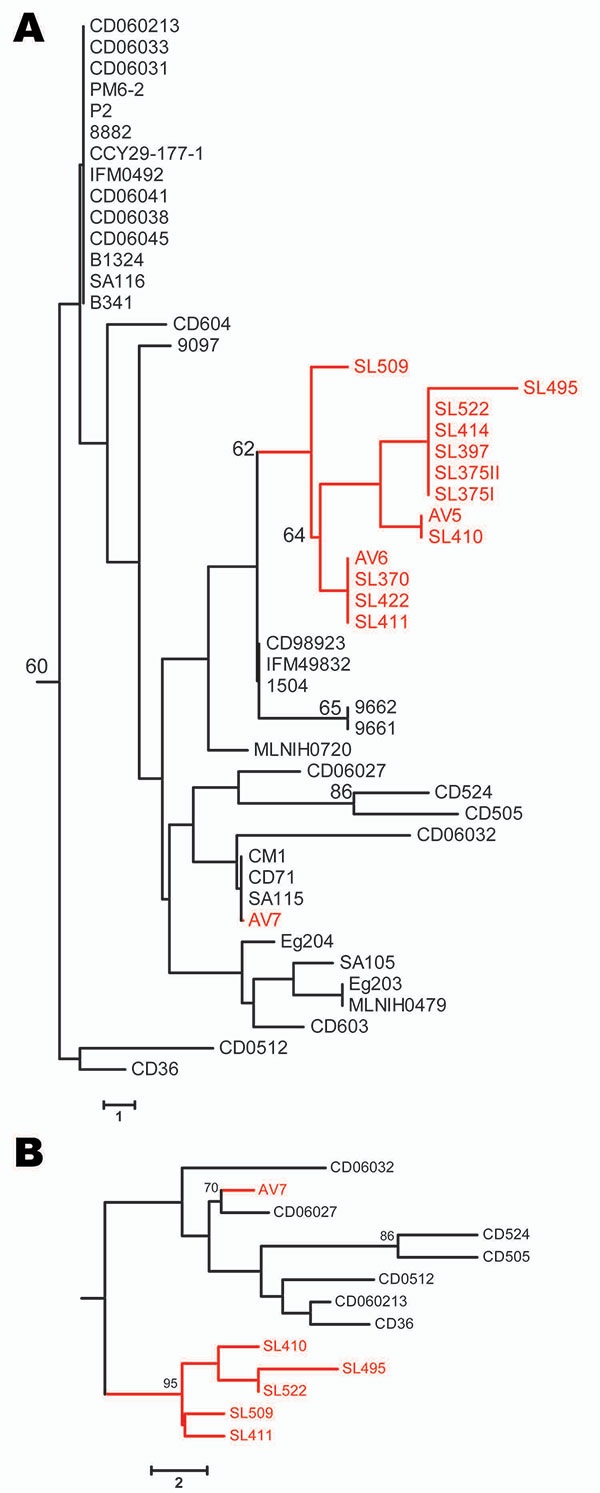
Figure. Neighbor-joining trees based on the polymorphic sites in Candida dubliniensis multilocus sequence typing (MLST) sequences. Bootstrap values >60% are indicated at cluster nodes. Avian-associated isolates are indicated in red. Numbers of polymorphic sites in isolates are indicated by scale bars. A) Isolates of MLST clade C1 defined by McManus et al. (7) showing location of avian-associated isolates in relation to human isolates in the same clade; human isolates were originally obtained in many countries. B) Neighbor-joining tree based on polymorphic sites in MLST sequences for each of 13 internal transcribed spacer genotype 1 C. dubliniensis isolates, 7 of which were obtained from humans in Ireland and 6 from seabird excrement in Ireland. Isolates that had identical diploid sequence types (DSTs) were not included in the tree so that only 1 of each DST is included. Tree displays the robustness of the avian-associated subgroup of isolates within a population of similar human-associated isolates from the same region. The rate of heterozygosity among human and avian-associated clade C1 isolates was 1.6 and 1 heterozygous site per DST, respectively, from 36 polymorphic sites, which indicated that avian-associated isolates were more clonal.
References
- Buck JD. Isolation of Candida albicans and halophilic Vibrio spp. from aquatic birds in Connecticut and Florida. Appl Environ Microbiol. 1990;56:826–8.PubMedGoogle Scholar
- Edelmann A, Kruger M, Schmid J. Genetic relationship between human and animal isolates of Candida albicans. J Clin Microbiol. 2005;43:6164–6. DOIPubMedGoogle Scholar
- Wrobel L, Whittington JK, Pujol C, Oh SH, Ruiz MO, Pfaller MA, Molecular phylogenetic analysis of a geographically and temporally matched collection of Candida albicans isolates from humans and nonmigratory wildlife in central Illinois. Eukaryot Cell. 2008;7:1475–86. DOIPubMedGoogle Scholar
- Jacobsen MD, Bougnoux ME, d’Enfert C, Odds FC. Multilocus sequence typing of Candida albicans isolates from animals. Res Microbiol. 2008;159:436–40. DOIPubMedGoogle Scholar
- Nunn MA, Schaefer SM, Petrou MA, Brown JR. Environmental source of Candida dubliniensis. Emerg Infect Dis. 2007;13:747–50.PubMedGoogle Scholar
- Odds FC, Jacobsen MD. Multilocus sequence typing of pathogenic Candida species. Eukaryot Cell. 2008;7:1075–84. DOIPubMedGoogle Scholar
- McManus BA, Coleman DC, Moran G, Pinjon E, Diogo D, Bougnoux ME, Multilocus sequence typing reveals that the population structure of Candida dubliniensis is significantly less divergent than that of Candida albicans. J Clin Microbiol. 2008;46:652–64. DOIPubMedGoogle Scholar
- Moran G, Sullivan D, Morschhauser J, Coleman D. The Candida dubliniensis CdCDR1 gene is not essential for fluconazole resistance. Antimicrob Agents Chemother. 2002;46:2829–41. DOIPubMedGoogle Scholar
- Coleman D, Sullivan D, Harrington B, Haynes K, Henman M, Shanley D, Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV-infected and AIDS patients. Oral Dis. 1997;3(Suppl 1):S96–101.PubMedGoogle Scholar
- Al Mosaid A, Sullivan DJ, Coleman DC. Differentiation of Candida dubliniensis from Candida albicans on Pal’s agar. J Clin Microbiol. 2003;41:4787–9. DOIPubMedGoogle Scholar
- Pincus DH, Coleman DC, Pruitt WR, Padhye AA, Salkin IF, Geimer M, Rapid identification of Candida dubliniensis with commercial yeast identification systems. J Clin Microbiol. 1999;37:3533–9.PubMedGoogle Scholar
- Donnelly SM, Sullivan DJ, Shanley DB, Coleman DC. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology. 1999;145:1871–82. DOIPubMedGoogle Scholar
- Gee SF, Joly S, Soll DR, Meis JF, Verweij PE, Polacheck I, Identification of four distinct genotypes of Candida dubliniensis and detection of microevolution in vitro and in vivo. J Clin Microbiol. 2002;40:556–74. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. DOIPubMedGoogle Scholar
- Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell. 2004;3:1015–27. DOIPubMedGoogle Scholar