Rare Influenza A (H3N2) Variants with Reduced Sensitivity to Antiviral Drugs
Clyde Dapat
1
, Yasushi Suzuki
1, Reiko Saito, Yadanar Kyaw, Yi Yi Myint, Nay Lin, Htun Naing Oo, Khin Yi Oo, Ne Win, Makoto Naito, Go Hasegawa, Isolde C. Dapat, Hassan Zaraket, Tatiana Baranovich, Makoto Nishikawa, Takehiko Saito, and Hiroshi Suzuki
Author affiliations: Niigata University, Niigata, Japan (C. Dapat, Y. Suzuki, R. Saito, M. Naito, G. Hasegawa, I.C. Dapat, H. Zaraket, T. Baranovich, H. Suzuki); National Institute of Animal Health, Tsukuba City, Japan (T. Saito); Niigata Prefectural Institute of Public Health and Environmental Sciences, Niigata (M. Nishikawa); Sanpya Hospital, Yangon, Myanmar (Y. Kyaw); National Health Laboratory, Yangon (K.Y. Oo, N. Win); Central Myanmar Department of Medical Research, Nay Pyi Taw, Myanmar (Y.Y. Myint, N. Lin, H.N. Oo)
Main Article
Figure 1
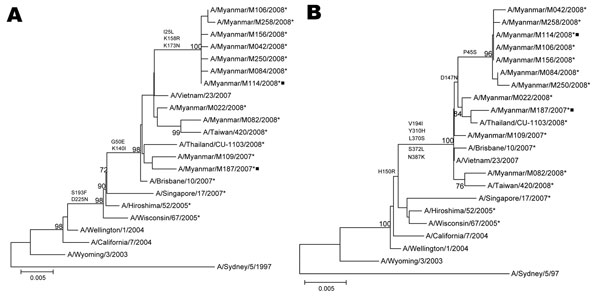
Figure 1. Phylogenetic analysis of the hemagglutinin (HA) (A) and neuraminidase (B) genes of influenza virus A (H3N2) isolates in Myanmar in 2007 and 2008. Trees were generated by using the neighbor-joining method. Bootstrap values >70% of 1,000 replicates and amino acid changes that characterize a branch are indicated on the left side of the node. Amantadine-resistant isolates with S31N mutation in M2 are marked with asterisks, and isolates with reduced sensitivity to zanamivir with Q136K mutation in NA are marked with squares. GenBank accession no. of the genomic sequences of isolates are GQ478849–GQ478866. Nucleotide sequences of the HA and NA genes of vaccine strains and isolates from other countries were obtained from the National Center for Biotechnology Information Influenza Virus Resource (www.ncbi.nlm.nih.gov/genomes/FLU). Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: December 14, 2010
Page updated: December 14, 2010
Page reviewed: December 14, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
