Novel Corynebacterium diphtheriae in Domestic Cats
Aron J. Hall

, Pamela K. Cassiday, Kathryn A. Bernard, Frances Bolt, Arnold G. Steigerwalt, Danae Bixler, Lucia C. Pawloski, Anne M. Whitney, Masaaki Iwaki, Adam Baldwin, Christopher G. Dowson, Takako Komiya, Motohide Takahashi, Hans P. Hinrikson, and Maria L. Tondella
Author affiliations: Centers for Disease Control and Prevention, Atlanta, Georgia, USA (A.J. Hall, P.K. Cassiday, A.G. Steigerwalt, L.C. Pawloski, A.M. Whitney, H.P. Henrikson, M.L. Tondella); West Virginia Department of Health and Human Resources, Charleston, West Virginia, USA (A.J. Hall, D. Bixler); Public Health Agency of Canada, Winnipeg, Manitoba, Canada (K.A. Bernard); University of Warwick, Coventry, UK (F. Bolt, A. Baldwin, C.G. Dowson); National Institute of Infectious Diseases, Tokyo, Japan (M. Iwaki, T. Komiya, M. Takahashi)
Main Article
Figure 1
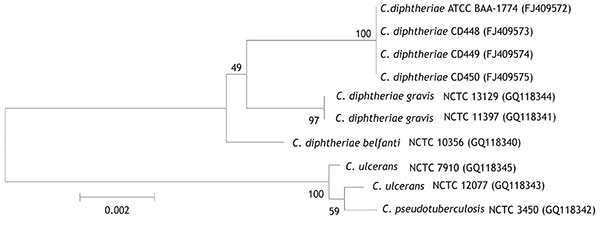
Figure 1. Neighbor-joining phylogenetic tree based on 16S rRNA gene sequence analysis of Corynebacterium diphtheriae isolates, including 4 feline isolates from West Virginia, 2008 (ATCC BAA-1774, CD 448, CD 449, CD 450). The tree was constructed from a 1,437-bp alignment of 16S rRNA gene sequences by using the neighbor-joining method and Kimura 2-parameter substitution model. Bootstrap values (expressed as percentages of 1,000 replicates) >40% are illustrated at branch points. Feline isolates had 100% identity with each other and >99.1% identity with C. diphtheriae biotypes gravis and belfanti. GenBank accession nos. given in parentheses. ATCC, American Type Culture Collection; CD, Centers for Disease Control and Prevention identifier number; NCTC, National Collection of Type Cultures. Scale bar indicates number of substitutions per site.
Main Article
Page created: December 28, 2010
Page updated: December 28, 2010
Page reviewed: December 28, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
