Volume 16, Number 5—May 2010
Letter
Triatoma infestans Bugs in Southern Patagonia, Argentina
Figure
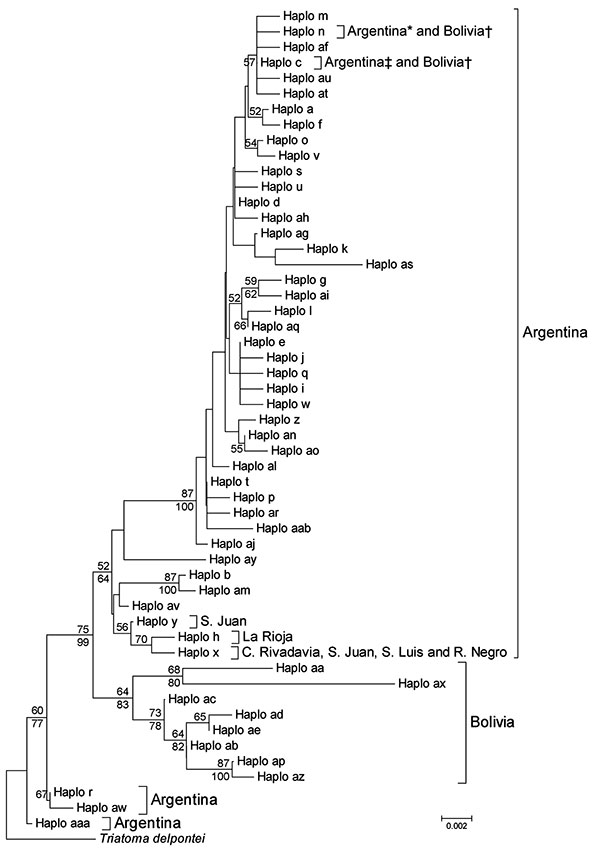
Figure. Phylogenetic relationships between mitochondrial cytochrome oxidase I gene haplotypes of Triatoma infestans from Argentina and Bolivia. The neighbor-joining tree was constructed by using MEGA 4.1 (www.megasoftware.net) and bootstrap values (based on 1,000 replications) >50% are shown above the branches. A Bayesian maximum clade credibility tree was similar, and clade posterior probabilities >50% are shown below the branches of the neighbor-joining tree. MRBAYES 3.1 (http://mrbayes.csit.fsu.edu) default priors were assumed and run for 4 million generations. Convergence of the Markov chain Monte Carlo analysis was investigated with the SD of split frequencies and diagnostics implemented in AWTY (http://ceb.csit.fsu.edu/awty). The model of evolution (Hasegawa-Kishino-Yano + invariable sites+ Γ [HKY + I + Γ]) was chosen with Mrmodeltest 2.3 (www.abc.se/~nylander). Because MEGA 4.1 does not support HKY; the more inclusive Tamura-Nei method (www.megasoftware.net/WebHelp/part_iv___evolutionary_analysis/computing_evolutionary_distances/distance_models/nucleotide_substitution_models/hc_tamura_nei_distance.htm) was used for the neighbor-joining analysis. Haplotypes al, an, ao, ap, aq, at, au, ax, az, aaa, and aab are reported. DNA sequences are available in GenBank (accession nos. EF451005-EF451041, FJ439768, FJ811845–8, and GQ 478993–GQ 479005). *Two provinces in Argentina; †Tarija, Bolivia; ‡10 provinces in Argentina. Scale bar indicates nucleotide substitutions per site.