Volume 10, Number 10—October 2004
Dispatch
Emerging Enteropathogenic Escherichia coli Strains?
Cite This Article
Citation for Media
Abstract
Escherichia coli strains of nonenteropathogenic serogroups carrying eae but lacking the enteropathogenic E. coli adherence factor plasmid and Shiga toxin DNA probe sequences were isolated from patients (children, adults, and AIDS patients) with and without diarrhea in Brazil. Although diverse in phenotype and genotype, some strains are potentially diarrheagenic.
Typical and atypical enteropathogenic Escherichia coli (EPEC) strains constitute two distinct groups of organisms that have in common the locus of enterocyte effacement (LEE), a pathogenicity island that promotes the development of attaching and effacing lesions (1,2). The LEE island encompasses the eae gene that encodes intimin, an outer membrane adhesin fundamental to the establishment of attaching and effacing lesions (1). Only typical EPEC strains bear the EPEC adherence factor (EAF) plasmid, in which a cryptic sequence used as a probe (EAF probe) to the category is located (1).
Various evidence suggests that atypical EPEC are closer to Shiga toxin–producing E. coli (STEC) (1), which cause diarrhea and hemolytic uremic syndrome (2). Although many STEC strains carry LEE, their main virulence mechanism is Shiga toxin(s) (Stx) production (2).
Twelve EPEC serogroups (O26, O55, O86, O111, O114, O119, O125, O126, O127, O128, O142, and O158) are recognized, but recent studies have shown that most typical EPEC strains fall into only certain O:H serotypes within these serogroups, which differ from those of atypical EPEC (1). Furthermore, E. coli strains of non-EPEC serogroups that carry eae but lack the EAF probe sequence and stx genes (eae+ EAF– stx– E. coli) have been detected (3–6), but their role in endemic diarrhea has not been established, and no precise understanding of them exists. Recently, we extensively characterized a collection of such strains from a single city in Brazil (6). To extend our knowledge on the diversity of eae+ EAF stx– E. coli strains of non-EPEC serogroups, we compared their occurrence in three distinct cities in Brazil and their genotypic and phenotypic characteristics.
The strains we studied were collected from patients of low socioeconomic status in three cities: São Paulo and Ribeirão Preto, in São Paulo State, and Rio de Janeiro, in Rio de Janeiro State, Brazil. The São Paulo strains were collected from 505 diarrheic and 505 nondiarrheic children (1–4 years of age) who visited Hospital Infantil Menino Jesus (April 1989–March 1990) (7). These strains had been previously characterized for various traits (6); in the present study, we tested them for new gene sequences. The Rio de Janeiro strains were collected from 372 diarrheic and 74 nondiarrheic children <5 years of age at the Instituto de Puericultura e Pediatria Martagão Gesteira, a public hospital at the Federal University of Rio de Janeiro (January 1998–December 1999, and May–December 2001). Strains from Ribeirão Preto were derived from 294 diarrheic children (<9 years of age) and adults (1–52 years), including 42 adults with AIDS. Fecal samples from these patients were sent to the Regional Laboratory of Instituto Adolfo Lutz by Hospital Santa Lydia and different clinics in the vicinity (August 2000–June 2002). This study has been approved by the Universidade Federal de São Paulo, Escola Paulista de Medicina Ethical Committee for human experimentation.
In all studies, five lactose-fermenting isolates and one nonlactose-fermenting isolate of each morphologic type, present in each fecal sample, were biochemically characterized as E. coli. Other well-established bacterial enteropathogens (Salmonella spp., Shigella spp., Aeromonas spp, Campylobacter spp., and Yersinia enterocolitica) and rotavirus were also searched for by standard methods (8).
All E. coli isolates were tested by colony hybridization with cloned or amplified genetic sequences for enterotoxigenic E. coli, enteroinvasive E. coli, EPEC (eae and EAF probes), STEC (stx probes), and enteroaggregative E. coli, as previously described (6). The E. coli strains that were eae+ EAF– stx– were serotyped at the Instituto Adolfo Lutz (National Reference Center for E. coli Serotyping) by using antisera O1 to O173 and H1 to H56.
In São Paulo and Rio de Janeiro, the eae+ EAF- stx- E. coli strains of non-EPEC serogroups occurred in similar frequencies in diarrheic and nondiarrheic children: 32 (6.3%) compared with 27 (5.3%), and 19 (5.1%) compared with 4 (5.4%), respectively. In Ribeirão Preto, such strains were found in 17 (5.8%) patients: 13 from children (1 month–9 years of age) and 4 from adults with AIDS (27–52 years of age). A total of 99 strains (one from each patient) were selected for further analysis. These strains had diverse serotypes (Table 1); 25 (25.2%) strains were nonmotile, 3 were rough, and 47 (47.5%) did not react with the O antisera tested. Among the 49 O-typable strains, 29 serogroups and 35 serotypes were found. The most frequent serotype was O51:H40 (10.1%), which occurred in all three areas studied. Most of the other serotypes occurred in one or two strains.
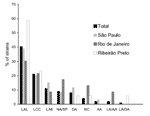
All strains were tested for adherence to HeLa cells (3- and 6-hour assays) (9). Four of them promoted sporadic adherence, four were nonadherent, and one was cytodetaching. For 88 of the 90 adherent strains, the adherence patterns could only be determined in 6 hours. Seventy-two (80.0%) of the 90 strains had variations of the localized adherence (LA) pattern of typical EPEC, which is characterized by compact bacterial clusters (10). These variant patterns included the following: LA-like pattern, which showed loose bacterial clusters (11); a pattern that showed loose and compact clusters; and a pattern identical to LA, despite its detection in only 6 hours (LA6). Other less frequent patterns included the following: the diffuse adherence typical of diffusely adhering E. coli, the aggregative adherence typical of enteroaggregative E. coli (2), and an association of diffuse adherence and LA or of aggregative adherence and LA. These mixed patterns were retained when individual colonies were tested. The aggregative adherence/LA pattern (two strains) was only recognized in the 3-hour assays. The prevalence of the different patterns varied by area of study, but the variations of LA were the most prevalent in all (72.7%) (Figure 1).
The ability to promote attaching and effacing lesions was tested by the fluorescent actin staining test (FAS) (7) in 94 strains; the 5 nonadherent or cytodetaching strains were not tested. Seventy (74.4%) of the strains tested were positive: 43 (72.9%), 15 (65.2%), and 12 (70.2%) of the strains from São Paulo, Rio de Janeiro, and Ribeirão Preto, respectively. Moreover, four distinct segments of the LEE region were found in all strains studied, as detected by hybridization with specific LEE sequences (LEE A, B, C, and D) (12), which suggests that all bear a complete LEE region.
LEE insertion sites were detected by a combination of polymerase chain reaction (PCR) assays with primers for the selC junctions and for conserved sequences of selC and pheU (12,13). LEE was inserted in selC in 46 strains: 24 (40.7%), 13 (56.6%), and 9 (53.0%) strains from São Paulo, Rio de Janeiro, and Ribeirão Preto, respectively. In addition, LEE was probably inserted in pheU in 29 (49.1%) and 3 (13.0%) of the São Paulo and Rio de Janeiro strains, respectively. In 13 strains, LEE is probably inserted in another site, since both loci were intact. The LEE insertion site was undetermined in eight strains because both selC and pheU were disrupted, and the primers for the LEE junctions in selC yield no amplification. Strains with an undetermined LEE insertion site occurred in all three areas studied.
Strains were also tested for 24 DNA sequences of established or putative virulence properties of pathogenic E. coli by colony hybridization (6). DNA probes were obtained from cloned genes (bfpA, perA, E-hly, EAEC, daaC, cdt, cnf, hly) (6) or by PCR amplification, which used as templates the genomic DNA of EAEC prototype strains 042 (aafC, aggR, aspU, shf, irp2, pet, and pic) and 17-2 (aggC and astA); extraintestinal pathogenic strains (ExPEC) J96 (pap) and KS52 (afa), and E. coli HB101 (pANN 801-13) (carrying the sfa probe). PCR primers and assay conditions used were described previously (6,14).
Hybridization with 17 of the 24 sequences tested was detected among the strains; hly and irp2 (31.3% each) and astA (29.3%) were the most frequent. Thirty-four different combinations of these 17 sequences were found (Table 2). Their prevalence varied by location, but 25 (73.5%) occurred in two or fewer strains. Among the less frequent combinations found, some were of genes of ExPEC and EAEC, and others of genes of EPEC (bfpA) and EHEC (E-hly). Moreover, 30.3% of the strains lacked all 24 DNA sequences tested, comprising the most frequent subgroup of strains in all three areas (Table 2). Although these strains carried only the eae gene and the four LEE probe sequences (LEE+ only strains), they may have carried virulence sequences other than those tested. Thus, one should not emphasize the virulence potential of these LEE+ strains solely on the basis of findings of significant differences in their frequencies between cases and controls.
DNA sequences similar to bfpA were detected in 14 (14.1%) of the 99 strains studied, however, only 2 expressed Bfp in Western blot experiments (not shown); these two strains also carried perA and presented AA/LA in 3 hours. The HeLa pattern of the remaining bfpA + strains varied, but none of them had compact clusters in 3 hours, which is typical of LA. Thus, Bfp expression was found only in strains presenting aggregative adherence/LA in 3 hours, as in typical LA of EPEC (1).
PCR assays with specific primers for the variable region of intimin were used to identify five intimin types (α, β, γ, δ, and ε) (15,16). Most strains had a nontypable intimin (64.6%), but the distribution of these strains varied (approximately 70% in São Paulo and 29%–35% in Rio de Janeiro and Ribeirão Preto). Recently, new schemes were proposed to identify intimin subtypes, which were not tested (17,18). The prevalence of typable intimins varied among the three areas analyzed. Intimin subtypes β (11.1%) and γ (12.1%) prevailed, and intimin ε was not found (Figure 2). The intimin types of two strains were not determined because amplification products of the expected size were obtained with four intimin pairs of primers.
In this study, we sought to verify the frequency with which eae+ EAF– stx– E. coli strains of non-EPEC serogroups occur in persons of poor socioeconomic status in three Brazilian cities; we also compared these strains’ genotypic and phenotypic characteristics. Although these strains occurred in 5% to 6% of the populations studied, including nondiarrheic children (in São Paulo and Ribeirão Preto), 73%–88 % of them were dissociated from other well-established enteropathogens (not shown).
Although O51:H40 was the most frequent serotype found and occurred in all three areas studied, the non-EPEC eae+ EAF– st– strains comprised a large variety of serotypes, and many were O nontypable. Moreover, the strains had diverse adherence patterns and various combinations of pathogenic E. coli DNA virulence sequences; the prevalence of these properties varied among the areas studied. Altogether, these data show that eae+ EAF– stx– E. coli strains outside the EPEC serogroups are even more diverse than already observed (6). As we have emphasized previously, such diversity challenges the diagnosis of these putative pathogens (6).
All strains carried an apparently complete LEE region, and approximately 75.0% of them had the potential to promote attaching and effacing lesions in HeLa cells, as detected by FAS. Thus at least these FAS+ strains are potentially enteropathogenic, since they are capable of inducing attaching and effacing lesions in vitro and may occur in diarrheic patients of various ages and in patients with AIDS. In the EPEC meeting held in 1995, a consensus definition of atypical EPEC was established, namely, that they are EAF–, eae+ strains that promote attaching and effacing lesions (19). Therefore, the FAS+ strains of our study could be classified as atypical EPEC. Whether these strains have additional virulence properties not present in typical EPEC remains to be elucidated. Studies on the virulence potential of selected strains at the cellular and molecular levels will certainly contribute to further understanding of this group of strains and aid in discriminating enteropathogenic strains within the group.
Dr. Gomes is associate professor of microbiology in the Department of Microbiology, Immunology and Parasitology at São Paulo Medical School, Federal University of São Paulo. She studies the epidemiology of diarrhea and the potential virulence factors of diarrheagenic E. coli, with emphasis on enteropathogenic, attaching-effacing, and enteroaggregative categories.
Acknowledgment
These studies were supported by FINEP/MCT/PRONEX grant (41.96.0881.00) and FAPESP grant 95/9176-4 awarded to T.A.T.G. and 01/07921-7 awarded to B.E.C.G.
References
- Trabulsi LR, Keller R, Gomes TAT. Typical and atypical enteropathogenic Escherichia coli (EPEC). Emerg Infect Dis. 2002;8:508–13.PubMedGoogle Scholar
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. [Erratum in Clin Microbiol Rev. 1998;11:403]. Clin Microbiol Rev. 1998;11:142–201.PubMedGoogle Scholar
- Afset JE, Bergh K, Bevanger L. High prevalence of atypical enteropathogenic Escherichia coli (EPEC) in Norwegian children with diarrhea. J Med Microbiol. 2003;52:1015–9. DOIPubMedGoogle Scholar
- Dulguer MV, Fabbricotti SH, Bando SY, Moreira-Filho CA, Fagundes-Neto U, Scaletsky ICA. Atypical enteropathogenic Escherichia coli strains: phenotypic and genotypic profiling reveals a strong association between enteroaggregative E. coli heat-stable enterotoxin and diarrhea. J Infect Dis. 2003;188:1685–94. DOIPubMedGoogle Scholar
- Regua-Mangia AH, Gomes TAT, Vieira MAM, Andrade JCR, Irino K, Teixeira LM. Frequency and characteristics of diarrheagenic Escherichia coli strains isolated from children with and without diarrhea in Rio de Janeiro, Brazil. J Infect. 2004;48:161–7. DOIPubMedGoogle Scholar
- Vieira MAM, Andrade JRC, Trabulsi LR, Rosa ACP, Dias AMG, Ramos SRTS, Phenotypic and genotypic characteristics of Escherichia coli strains of non-enteropathogenic E. coli (EPEC) serogroups that carry eae and lack the EPEC adherence factor and Shiga toxin DNA probe sequences. J Infect Dis. 2001;183:762–72. DOIPubMedGoogle Scholar
- Gomes TAT, Griffin PM, Ivey C, Trabulsi LR, Ramos SRTS. EPEC Infections in São Paulo. Rev Microbiol, São Paulo. 1996;27:25–33.
- Cravioto A, Gross RJ, Scotland SM, Rowe B. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol. 1979;3:95–9. DOIGoogle Scholar
- Scaletsky ICA, Silva MLM, Trabulsi LR. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45:534–6.PubMedGoogle Scholar
- Rodrigues J, Scaletsky ICA, Campos LC, Gomes TAT, Whittam ST, Trabulsi LR. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect Immun. 1996;64:2680–6.PubMedGoogle Scholar
- Knutton S, Baldwin T, Williams PH, McNeish AS. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–13.PubMedGoogle Scholar
- McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–8. DOIPubMedGoogle Scholar
- Sperandio V, Kaper JB, Bortolini MR, Neves BC, Keller R, Trabulsi LR. Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol Lett. 1998;164:133–9. DOIPubMedGoogle Scholar
- Elias WP, Uber AP, Tomita S, Trabulsi LR, Gomes TAT. Combinations of putative virulence markers in typical and variant enteroaggregative Escherichia coli strains from children with and without diarrhea. Epidemiol Infect. 2002;129:49–55. DOIPubMedGoogle Scholar
- Adu-Bobie J, Frankel G, Bain C, Gonçalves AG, Trabulsi LR, Douce G, Detection of intimin alpha, beta, gamma, and delta, four intimin derivatives by attaching and effacing microbial pathogens. J Clin Microbiol. 1998;36:662–8.PubMedGoogle Scholar
- Oswald E, Schmidt H, Morabito S, Karch H, Marchès O, Caprioli A. Typing of intimin genes in human and animal enterohemorrhagic Escherichia coli: characterization of a new intimin variant. Infect Immun. 2000;68:64–71. DOIPubMedGoogle Scholar
- Jenkins C, Lawson AJ, Cheasty T, Willshaw GA, Wright P, Dougan G, Subtyping intimin genes from enteropathogenic Escherichia coli associated with outbreaks and sporadic cases in the United Kingdom and Eire. Mol Cell Probes. 2003;17:149–56. DOIPubMedGoogle Scholar
- Zhang WL, Köhler B, Oswald E, Beutin L, Karch H, Morabito S, Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J Clin Microbiol. 2002;40:4486–92. DOIPubMedGoogle Scholar
- Kaper JB. Defining EPEC. Proceedings of the International Symposium on Enteropathogenic Escherichia coli (EPEC). Rev Microbiol São Paulo. 1996;27:130–3.
Figures
Tables
Cite This ArticleTable of Contents – Volume 10, Number 10—October 2004
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Address for corresponden ce: Tânia A. Tardelli Gomes, Departamento de Microbiologia, Imunologia e Parasitologia, Universidade Federal de São Paulo, Escola Paulista de Medicina, Rua Botucatu, 862, 3°andar, Vila Clementino, São Paulo, São Paulo, Brazil, CEP 04023-062; fax: 55-11-5571-6504
Top