Volume 11, Number 11—November 2005
Research
Cryptococcus gattii in AIDS Patients, Southern California
Figure 1
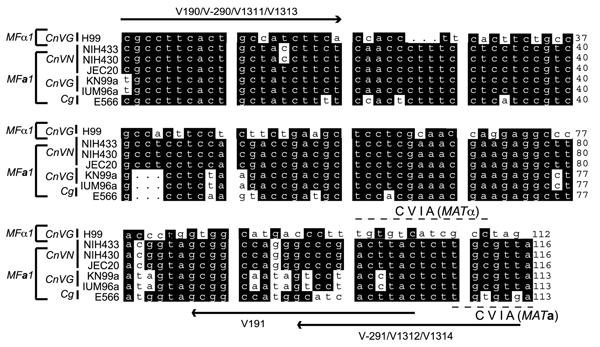
Figure 1. Primers for pheromone polymerase chain reaction (PCR). Nucleotide sequence alignment for MFα1 and MFa1 genes is shown with characteristic Cys-Val-Ile-Ala (CVIA) motifs. Both sense and antisense primers were designed from within the open reading frames of pheromone genes, to ensure high specificity of the multiplex PCR. The MFα1 sequence from Cryptococcus neoformans var. grubii (CnVG) (AF542529) and the MFa1 sequences from CnVG (AY129299), Cryptococcus neoformans var. neoformans (CnVN) (AF542530), and Cryptococcus gattii (Cg) (AY710429) were used for multiple alignments with GCG (Wisconsin package version 10.0). A common primer pair, V190/V191, was designed to get MFα1 PCR amplicons from CnVG, CnVN, and Cg (MFα1 sequence from CnVG was used as a reference), while unique primer pairs V290/V291, V1311/V1312, and V1313/V1314 were designed to get MFa1 PCR amplicons from CnVN, CnVG, and Cg, respectively. All the 3´-PCR primers contained a sequence from CVIA motif, which provided specificity to PCRs for pheromone genes.