Volume 11, Number 11—November 2005
Research
Cryptococcus gattii in AIDS Patients, Southern California
Figure 4
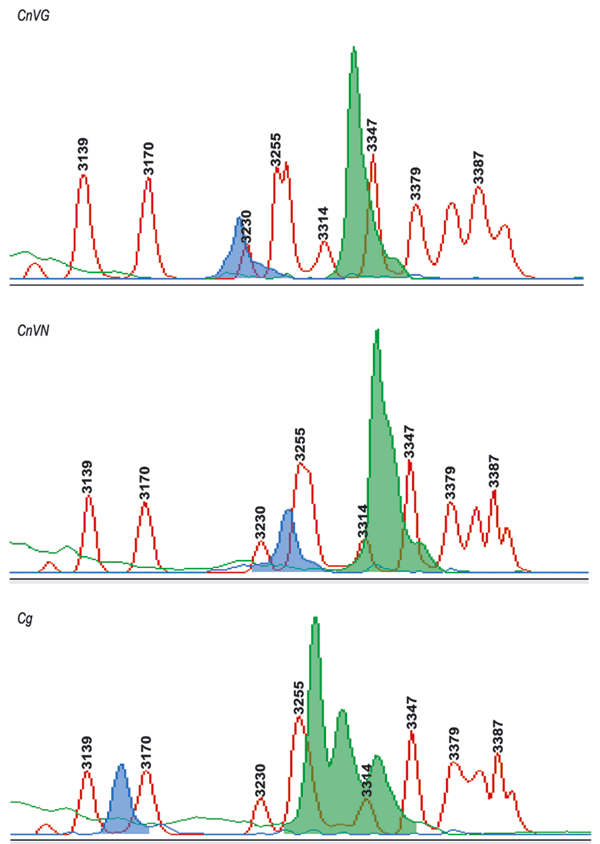
Figure 4. Capillary electrophoresis–single strand conformation polymorphisms (CE-SSCP) for the identification of varieties and species. The ABI PRISM 310 Genetic Analyzer and GeneScan analysis software were used for variety and species determination with the MFα1 pheromone gene. The MFα1 sense and antisense primers were labeled with fluorescent probes FAM (blue) and TET (green), and polymerase chain reaction amplicons were analyzed with 3% polymer at 30°C under nondenaturing conditions. The blue and green peaks depict characteristic peak pattern for Cryptococcus neoformans var. grubii (CnVG), Cryptococcus neoformans var. neoformans (CnVN), and Cryptococcus gattii (Cg). These peaks were aligned by using an internal size standard.
Page created: February 17, 2012
Page updated: February 17, 2012
Page reviewed: February 17, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.