Volume 11, Number 5—May 2005
Dispatch
Human T-cell Leukemia Virus Type 1 Molecular Variants, Vanuatu, Melanesia
Figure 2
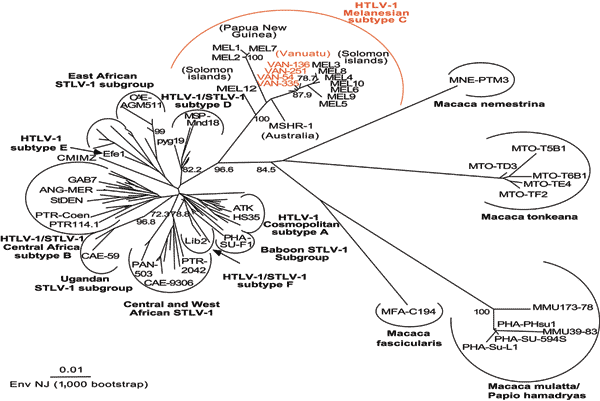
Figure 2. . Unrooted phylogenetic tree generated by the neighbor-joining method by using the 522-bp fragment of the env gene. Distance matrices were generated with the DNADIST program, using the Kimura 2-parameter method and 5.65 as the transition/transversion ratio. Bootstrap analysis was carried out with 1,000 datasets. The values on the branches indicate frequencies of occurrence for 1,000 trees. The 4 new human T-cell leukemia virus type 1 (HTLV-1) sequences (VAN 54, VAN 136, VAN 251, VAN 335; GenBank accession nos. AY549879, AY549880, AY549881, AY549882) were analyzed with 126 HTLV-1/simian T-cell leukemia virus type 1 (STLV-1) sequences available from the GenBank database. The branch lengths are proportional to the evolutionary distance between the taxa.