Volume 11, Number 7—July 2005
Dispatch
Yersinia pseudotuberculosis Septicemia and HIV
Figure
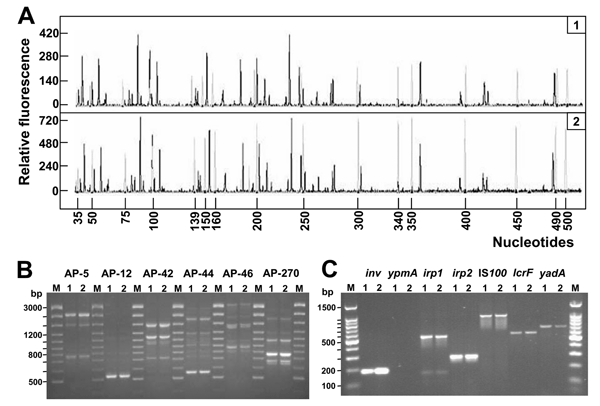
Figure. . Genetic fingerprinting and detection of virulence genes of Yersinia pseudotuberculosis isolates. A) Fluorescent amplified fragment length polymorphism (AFLP) analysis of Y. pseudotuberculosis DNA (black electropherogram; 1 and 2 refer to patient number). Reactions were performed as indicated in the AFLP Microbial Fingerprinting kit (Applied Biosystems, Foster City, CA, USA). Reference DNA from Escherichia coli W3110 (Applied Biosystems) was used as internal control (gray electropherogram). Separation and detection of the AFLP fragments were performed with the Applied Biosystems model 3100 capillary electrophoresis system equipped with a 36-cm capillary loaded with the POP-4 polymer. Size determinations of the labeled DNA fragments were performed automatically with the Genescan Analysis 3.0 software (Applied Biosystems). B) Arbitrarily primed polymerase chain reaction (AP-PCR) analysis with a set of 6 oligonucleotides: AP5 (5´-TCCCGCTGCG-3´), AP12 (5´-CGGCCCCTGC-3´), AP42 (5´-AACGCGCAAC-3´), AP44 (5´-AGCCAGTTTC-3´), AP46 (5´-GAGGACAAAG-3´), and AP270 (5´-TGCGCGCGGG-3´) (8,9). Amplification patterns of DNA from the 2 clinical isolates are shown: lane 1, patient 1; lane 2, patient 2. M, molecular weight marker. The numbers on the left indicate the length (in base pairs) of the reference ladder. Primers are indicated on top. C) Detection of Y. pseudotuberculosis virulence genes. Primers and PCR conditions have been described elsewhere (2,6). Lane 1, patient 1; lane 2, patient 2. Lane M, molecular weight marker. The numbers on the left indicate the length (in base pairs) of the reference ladder. Target genes are indicated on top.