Volume 11, Number 7—July 2005
Research
Norovirus Recombination in ORF1/ORF2 Overlap
Figure 1
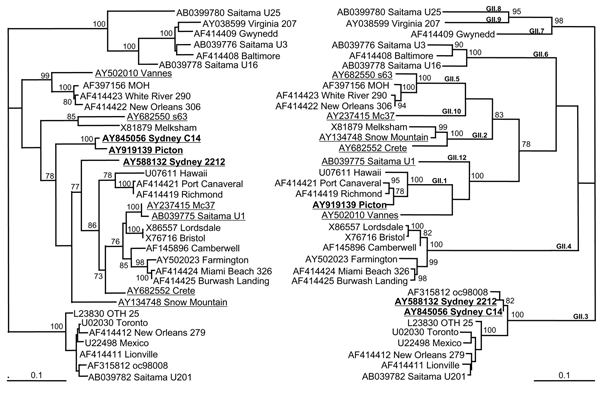
Figure 1. . Phylogenetic analysis of the nucleotide sequences of capsid and polymerase regions of 9 identified recombinant norovirus genogroup II strains in relation to 26 known strains and prototype strains. The left tree analyzes the relationship of a 420-bp region of the 3´ end of the polymerase region. The right tree shows the relationship of 550 bp of the 5´ end of the capsid sequence. Suspected recombinants are underlined to emphasize their different phylogenetic groupings, and strains described in this study are represented in boldface. The percentage bootstrap values in which the major groupings were observed among 100 replicates are indicated. The branch lengths are proportional to the evolutionary distance between sequences and the distance scale, in nucleotide substitutions per position, is shown. The capsid clustering is shown in bold and is based on the classification of Vinjé et al. (23) (Table).
References
- Koopmans MPG, von Bonsdorrf CH, Vinje J, DeMedici D, Monroe SS. Foodborne enteric viruses. FEMS Microbiol Rev. 2002;26:187–205.PubMedGoogle Scholar
- Fankhauser RL, Monroe SS, Noel JS, Humphrey CD, Bresee JS, Parashar UD, Epidemiological and molecular trends of "Norwalk-like viruses" associated with outbreaks of gastroenteritis in the United States. J Infect Dis. 2002;186:1–7. DOIPubMedGoogle Scholar
- Noel JS, Fankhauser RL, Ando T, Monroe SS, Glass RI. Identification of a distinct common strain of "Norwalk-like viruses" having a global distribution. J Infect Dis. 1999;179:1334–44. DOIPubMedGoogle Scholar
- Atmar RL, Estes MK. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin Microbiol Rev. 2001;14:15–37. DOIPubMedGoogle Scholar
- Green KY, Chanock RM, Kapikan AZ. Human calicivirus. In: Knipe DM, Howley PM, editors. Fields virology. Volume 1. 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 841–74.
- Bertolotti-Ciarlet A, White LJ, Chen R, Prasad BV, Estes MK. Structural requirements for the assembly of Norwalk virus-like particles. J Virol. 2002;76:4044–55. DOIPubMedGoogle Scholar
- Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80:2535–43.PubMedGoogle Scholar
- Lai MMC. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79.PubMedGoogle Scholar
- Hardy ME, Kramer SE, Treanor JJ, Estes MK. Human calicivirus genogroup II capsid sequence diversity revealed by analyses of the prototype Snow Mountain agent. Arch Virol. 1997;142:1469–79. DOIPubMedGoogle Scholar
- Katayama K, Shirato-Horikoshi H, Kojima S, Kageyama T, Oka T, Hoshino FB, Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology. 2002;299:225–39. DOIPubMedGoogle Scholar
- Hansman GS, Katayama K, Peerakome N, Khamrin P, Tonusin S, Okitsu S, Genetic diversity of norovirus and sapovirus in hospitalized infants with sporadic cases of acute gastroenteritis in Thailand. J Clin Microbiol. 2004;42:1305–7. DOIPubMedGoogle Scholar
- Jiang X, Espul C, Zhong WM, Cuello H, Matson DO. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch Virol. 1999;144:2377–87. DOIPubMedGoogle Scholar
- Hansman GS, Doan LTP, Kguyen TA, Okitsu S, Katayama K, Ogawa S, Detection of norovirus and sapovirus infection among children with gastroenteritis in Ho Chi Minh City, Vietnam. Arch Virol. 2004;149:1673–88. DOIPubMedGoogle Scholar
- Han MG, Smiley JR, Thomas C, Saif LJ. Genetic recombination between two genotypes of genogroup III bovine noroviruses (BoNVs) and capsid sequence diversity among BoNVs and Nebraska-like bovine enteric caliciviruses. J Clin Microbiol. 2004;42:5214–24. DOIPubMedGoogle Scholar
- Oliver SL, Brown DW, Green J, Bridger JC. A chimeric bovine enteric calicivirus: evidence for genomic recombination in genogroup III of the Norovirus genus of the Caliciviridae. Virology. 2004;326:231–9. DOIPubMedGoogle Scholar
- Katayama K, Miyoshi T, Uchino K, Oka T, Tanaka T, Naokazu T, Novel recombinant sapovirus. Emerg Infect Dis. 2004;10:1874–6.PubMedGoogle Scholar
- White PA, Hansman GS, Li A, Dable J, Isaacs M, Ferson M, Norwalk-like virus 95/96-US strain is a major cause of gastroenteritis outbreaks in Australia. J Med Virol. 2002;68:113–8. DOIPubMedGoogle Scholar
- Felsenstein J. PHYLIP inference package, version 3.5c. Seattle: University of Washington; 1993.
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8.PubMedGoogle Scholar
- Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–9. DOIPubMedGoogle Scholar
- Lole K, Bollinger R, Paranjape R, Gadkari D, Kulkarni S, Novak N, Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–60.PubMedGoogle Scholar
- Posada D. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol Biol Evol. 2002;19:708–17. DOIPubMedGoogle Scholar
- Vinjé J, Hamidjaja RA, Sobsey MD. Developmental and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J Virol Methods. 2004;116:109–17. DOIPubMedGoogle Scholar
- Vinjé J, Green J, Lewis DC, Gallimore CI, Brown DW, Koopmans MP. Genetic polymorphism across regions of the three open reading frames of "Norwalk-like viruses.". Arch Virol. 2000;145:223–41. DOIPubMedGoogle Scholar
- Gallimore CI, Green J, Richards AF, Cotterill H, Curry A, Brown DW, Methods for the detection and characterisation of noroviruses associated with outbreaks of gastroenteritis: outbreaks occurring in the north-west of England during two norovirus seasons. J Med Virol. 2004;73:280–8. DOIPubMedGoogle Scholar
- Meyers G, Wirblich C, Thiel HJ. Rabbit hemorrhagic disease virus—molecular cloning and nucleotide sequencing of a calicivirus genome. Virology. 1991;184:664–76. DOIPubMedGoogle Scholar
- Herbert TP, Brierley I, Brown TDK. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic RNA. J Gen Virol. 1996;77:123–7. DOIPubMedGoogle Scholar
- Pletneva MA, Sosnovtsev SV, Green KY. The genome of Hawaii virus and its relationship with other members of the Caliciviridae. Virus Genes. 2001;23:5–16. DOIPubMedGoogle Scholar
- Miller WA, Dreher TW, Hall TC. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (-) sense genomic RNA. Nature. 1985;313:68–70. DOIPubMedGoogle Scholar
- Morales M, Barcena J, Ramirez MA, Boga JA, Parra F, Torres JM. Synthesis in vitro of rabbit hemorrhagic disease virus subgenomic RNA by internal initiation on (-) sense genomic RNA. J Biol Chem. 2004;279:17013–8. DOIPubMedGoogle Scholar
- Kao CC, Singh P, Ecker DJ. De novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–60. DOIPubMedGoogle Scholar
- Cooper PD, Steiner-Pryor A, Scotti PD, Delong D. On the nature of poliovirus genetic recombinants. J Gen Virol. 1974;23:41–9. DOIPubMedGoogle Scholar
- Miller WA, Koev G. Minireview: synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. 2000;273:1–8. DOIPubMedGoogle Scholar
- Ishikawa M, Janda M, Krol MA, Ahlquist P. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J Virol. 1997;71:7781–90.PubMedGoogle Scholar
- Haseltine WA, Kleid DG, Panet A, Rothenberg E, Baltimore D. Ordered transcription of RNA tumor virus genomes. J Mol Biol. 1976;106:109–31. DOIPubMedGoogle Scholar
- van Marle G, Dobbe JC, Gultyaev AP, Luytjes W, Spaan WJ, Snijder EJ. Artevirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulation sequences. Proc Natl Acad Sci U S A. 1999;96:12056–61. DOIPubMedGoogle Scholar