Volume 11, Number 8—August 2005
Research
HIV-1 Genetic Diversity in Antenatal Cohort, Canada
Figure
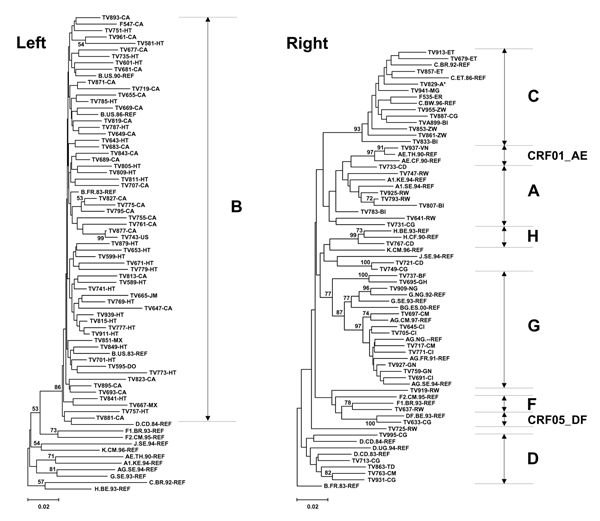
Figure. . Phylogenetic analysis of pol sequences derived from pregnant women infected with HIV-1. Trees were constructed by using the neighbor-joining method as described in Patients and Methods. A transition/transversion ratio of 2 was used and 1,000 bootstrap resamplings were performed. Left panel: Subgrouping with clade B HIV-1. Right panel: Grouping with non-B HIV. Reference sequences (REF) were obtained from the Los Alamos National Laboratory database (2001) (8). The scale bar represents 0.02 nucleotide substitutions per site. Letter codes indicate country of origin. All nucleotide sequence information was submitted to GenBank (accession no. DQ059647–DQ059749). CRF, circulating recombinant form.
References
- Thomson MM, Perez-Avarez L, Najera R. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect Dis. 2002;2:461–71. DOIPubMedGoogle Scholar
- Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin MC, Saragosti S, Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–7. DOIPubMedGoogle Scholar
- Peeters M, Sharp PM. Genetic diversity of HIV-1: the moving target. AIDS. 2000;14(Suppl 3):S129–40.PubMedGoogle Scholar
- Spira S, Wainberg MA, Loemba H, Turner D, Brenner BG. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J Antimicrob Chemother. 2003;51:229–40. DOIPubMedGoogle Scholar
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–60. DOIPubMedGoogle Scholar
- McCutchan FE. Understanding the genetic diversity of HIV-1. AIDS. 2000;14(Suppl 3):S31–44.PubMedGoogle Scholar
- Kuiken CL, Foley B, Freed E, Hahn B, Korber B, Marx PA, , eds. HIV sequence compendium 2002. Los Alamos (NM): Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 2002.
- Vallet S, Legrand-Quillien MC, Roger C, Bellein V, Perfezou P, de Saint-Martin L, HIV-1 genetic diversity in Western Brittany, France. FEMS Immunol Med Microbiol. 2002;34:65–71. DOIPubMedGoogle Scholar
- Fransen K, Buve A, Nkengasong JN, Laga M, van der Groen G. Longstanding presence in Belgians of multiple non-B HIV-1 subtypes. Lancet. 1996;347:1403. DOIPubMedGoogle Scholar
- Thomson MM, Delgado E, Manjon N, Ocampo A, Villahermosa ML, Marino A, HIV-1 genetic diversity in Galicia, Spain: BG intersubtype recombinant viruses are circulating among injecting drug users. AIDS. 2001;15:509–16. DOIPubMedGoogle Scholar
- Boni J, Pyra H, Gebhardt M, Perrin L, Burgisser P, Matter L, High frequency of non-B subtypes in newly diagnosed HIV-1 infections in Switzerland. J Acquir Immune Defic Syndr. 1999;22:174–9.PubMedGoogle Scholar
- Perrin L, Kaiser L, Yerly S. Travel and spread of HIV-1 genetic variants. Lancet Infect Dis. 2003;3:22–7. DOIPubMedGoogle Scholar
- Cuevas MT, Ruibal I, Villahermosa ML, Diaz H, Delgado E, Vasquez-de Parga E, High HIV-1 genetic diversity in Cuba. AIDS. 2002;16:1643–53. DOIPubMedGoogle Scholar
- Weidle PJ, Ganea CE, Irwin KL, Pieniazek D, McGowan JP, Oliva N, Presence of human immunodeficiency (HIV) type 1, group M, non-B subtypes, Bronx, New York: a sentinel site for monitoring HIV diversity in the United States. J Infect Dis. 2000;181:470–5. DOIPubMedGoogle Scholar
- Delwart EL, Orton S, Parekh B, Dobbs T, Clark K, Busch MP. Two percent of HIV-positive U.S. blood donors are infected with non-subtype B strains. AIDS Res Hum Retroviruses. 2003;19:1065–70. DOIPubMedGoogle Scholar
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;997:4876–82.PubMedGoogle Scholar
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. DOIPubMedGoogle Scholar
- Leitner T, Escanilla D, Franzen C, Uhlen M, Albert J. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc Natl Acad Sci U S A. 1996;93:10864–9. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–5. DOIPubMedGoogle Scholar
- Siepel AC, Korber BT. Scanning the database for recombinant HIV-1 genomes. In: Myers G, Korber B, Hahn BH, Jeang KT, Mellors JW, McCutchan FE, et al., editors. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos (NM): Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1995. p. III-35–III-60.
- Sullivan PS, Do AN, Ellenberger D, Pau CP, Paul S, Robbins K, Human immunodeficiency virus (HIV) subtype surveillance of African-born persons at risk for group O and group N HIV infections in the United States. J Infect Dis. 2000;181:463–9. DOIPubMedGoogle Scholar
- Eshleman SH, Hackett J, Swanson P, Cunningham SP, Drews B, Brennan C, Performance of the Celera Diagnostics ViroSeq HIV-1 Genotyping System for sequence-based analysis of diverse human immunodeficiency virus type 1 strains. J Clin Microbiol. 2004;42:2711–7. DOIPubMedGoogle Scholar
- Montavon C, Toure-Kane C, Liegois F, Mpoudi E, Bourgeois A, Vergne L, Most env and gag subtype A HIV-1 viruses circulating in west and west central Africa are similar to the prototype AG recombinant virus IBNG. J Acquir Immune Defic Syndr. 2000;23:363–74.PubMedGoogle Scholar
- Bikandou B, Takehisa J, Mboudjeka I, Ido E, Kuwata T, Miyazaki Y, Genetic subtypes of HIV type 1 in Republic of Congo. AIDS Res Hum Retroviruses. 2000;16:613–9. DOIPubMedGoogle Scholar
- Adrien A, Leaune V, Remis RS, Boivin JF, Rud E, Duperval R, Migration and HIV: an epidemiological study of Montrealers of Haitian origin. Int J STD AIDS. 1999;10:237–42. DOIPubMedGoogle Scholar
- Jayaraman GC, Gleeson T, Rekart ML, Cook D, Preiksaitis J, Sidaway F, Prevalence and determinants of HIV-1 subtypes in Canada: enhancing routinely collected information through the Canadian HIV Strain and Drug Resistance Surveillance Program. Can Commun Dis Rep. 2003;29:29–36.PubMedGoogle Scholar
- Brodine SK, Shaffer RA, Starkey MJ, Tasker SA, Gilcrest JL, Louder MK, Drug resistance patterns, genetic subtypes, clinical features, and risk factors in military personnel with HIV-1 seroconversion. Ann Intern Med. 1999;131:502–6.PubMedGoogle Scholar
- Brodine SK, Starkey MJ, Shaffer RA, Ito SI, Tasker SA, Barile AJ, Diverse HIV-1 subtypes and clinical, laboratory and behavioral factors in a recently infected US military cohort. AIDS. 2003;17:2521–7. DOIPubMedGoogle Scholar
- United Nations High Commissioner for Refugees. The Rwandan genocide and its aftermath. In: The state of the world's refugees 2000¾fifty years of humanitarian action. London: Oxford University Press; 2000, p. 245–88.
- Direction de la population et de la recherche. Tableaux sur l'immigration au Québec. Montreal: Ministère de l'immigration et des communautés culturelles; 2003. p. 15–32.
Page created: April 23, 2012
Page updated: April 23, 2012
Page reviewed: April 23, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.