Volume 12, Number 1—January 2006
Dispatch
Novel Parvovirus and Related Variant in Human Plasma
Figure 1
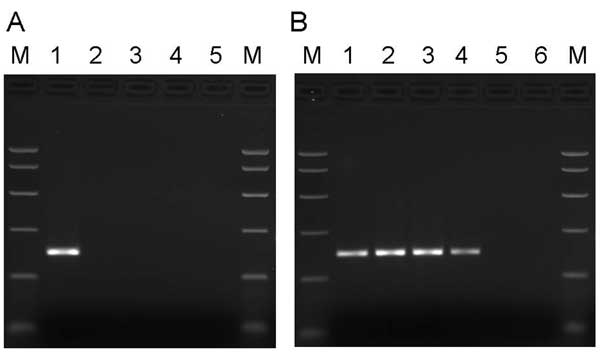
Figure 1. A) Specificity of primers for PARV4. Samples in lanes 1–5 were amplified by using primers directed to open reading frame 1 (ORF1) of PARV4. Template DNA in lane 1 was a plasmid subclone of the PARV4 ORF1 region. In lane 2, the template DNA was derived from parvovirus B19 International Standard (99/800, National Institute for Biological Standards and Control, South Mimms, UK) as representative of genotype 1 erythrovirus sequences; in lane 3, the template DNA was derived from a genotype 2 erythrovirus plasmid clone (A6; obtained from K. Brown, National Heart, Lung and Blood Institute, Bethesda, MD, USA); in lane 4, the template DNA was derived from a genotype 3 erythrovirus plasmid clone (D91.1; obtained from A. Garbarg-Chenon, Hôpital Trousseau, Paris, France). Template DNA in the erythrovirus samples (lanes 2–4) was adjusted to give ≈105.5 copies of each genotype per reaction. Lane 5, no template control. Polymerase chain reaction (PCR) products were analyzed on a 2.5% agarose gel alongside PCR Markers (M) (Promega, Madison, WI, USA). B) Screening manufacturing plasma samples for PARV4. Samples in lanes 1–6 were amplified by using primers directed to the ORF1 region of PARV4. Template DNA in lanes 1 and 2 consisted of 1 × 102 and 1 × 103 copies of the ORF1 subclone of PARV4. In lane 3, the template DNA was derived from a plasma pool containing 3.9 × 106 PARV4 genome copies/mL plasma; in lane 4, the template DNA was derived from a plasma pool containing <500 PARV4 genome copies/mL plasma; in lane 5, the template DNA was derived from a plasma pool that tested negative for PARV4 sequences. Lane 6, no template control. PCR products were analyzed on a 2.5% agarose gel alongside PCR Markers (M) (Promega).