Volume 12, Number 10—October 2006
Dispatch
Chikungunya Outbreaks Caused by African Genotype, India
Cite This Article
Citation for Media
Abstract
Chikungunya fever is reported in India after 32 years. Immunoglobulin M antibodies and virus isolation confirmed the cause. Phylogenic analysis based on partial sequences of NS4 and E1 genes showed that all earlier isolates (1963–1973) were Asian genotype, whereas the current and Yawat (2000) isolates were African genotype.
Chikungunya virus (CHIKV) outbreaks have been documented in Africa and Southeast Asia. In India, the first CHIKV outbreak was recorded in 1963 in Calcutta and was followed by epidemics in Chennai, Pondicherry, and Vellore in 1964; Visakhapatnam, Rajmundry, and Kakinada in1965; Nagpur in1965; and Barsi in 1973 (1). Recently, CHIKV has emerged in Southeast Asia and the Pacific region (2–4). Massive outbreaks have been reported from many islands in the Indian Ocean (5). Aedes albopictus is considered the vector in Reunion and other islands in the Indian Ocean (5), but Ae. aegypti is the main vector in Asia, including India (1). We investigated a large number of patients with fever with arthralgia, reported from October 2005 through March 2006, in many districts from Andhra Pradesh, Karnataka, and Maharashtra states.
Blood samples were collected from 1,938 suspected case-patients from the 3 states; serum was separated and transported to the laboratory on wet ice. Adult mosquitoes were collected from houses and sheds. Larval mosquitoes were collected from the affected areas by single-larva survey method. Adult household indexes and Breteau indexes were calculated for each area (6).
The C6/36 cell line was used for virus isolation (7). Immunoglobulin M (IgM) antibodies to CHIKV (IgM anti-CHIK) and dengue virus (IgM anti-dengue) (8) were assayed by IgM capture ELISA. For CHIKV ELISA, brain suspensions from mice infected with CHIKV were the source of antigen, and monoclonal antibodies were the source of antibodies (9). Dengue/CHIKV IgM antibodies and negative control human sera were included for respective tests. Approval for use of mice for antigen preparation was obtained from the institutional ethical committee according to national guidelines.
Immunofluorescence assay (IFA) was used to detect the virus in cell culture and in crushed heads of adult mosquitoes (10). A patient with the following was confirmed as having CHIKV infection: acute onset of moderate-to-high fever with joint pain of varying severity; negative test results for malaria, typhoid, and tuberculosis; and positive results for IgM anti-CHIKV antibodies, seroconversion, or CHIKV isolation. We used χ2 test to compare proportions of cases in different age groups.
We studied CHIKV isolates obtained during current investigations and viruses isolated during earlier epidemics in India (1963–2000) (Table 1). RNA was isolated by using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Superscript II (Invitrogen, Carlsbad, CA, USA) was used for reverse transcription (42°C for 1 h). Initially, Alphavirus genus–specific primers that produced a 472-bp fragment (NS4 gene) were F1 5´ GAY GCI TAY YTI GAY ATG GTI GAI GG 3´ and R1 5´ KYT CYT CIG TRT GYT TIG TIC CIGG 3´ (11). The second set of primers that amplified a 294-bp product of E1 gene were CHIK/E1-S 5´ TAC CCA TTC ATG TGG GGC 3´ and CHIK/E1-C 5´ GCC TTT GTA CAC CAC GATT 3´ (12). For amplification, Platinum Pfx enzyme (Invitrogen) was used. Cycling conditions were 1 cycle at 94°C for 5 min; then 35 cycles each of 94°C (1 min), 50°C (1 min), and 68°C (1.5 min); followed by final extension of 7 min at 68°C. The PCR products were purified by using QIAquick PCR Purification Kit (Qiagen) and sequenced by using BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) and an automatic sequencer (ABI PRISM 3100 Genetic Analyzer, Applied Biosystems).
Using ClustalX, version 1.83, multiple alignments of nucleotide sequences were performed. The phylogenic status of the CHIKV isolates was assessed with the software MEGA 3.1 (13), Kimura 2-parameter distance, and neighbor-joining algorithm. The reliability of different phylogenic groupings was evaluated with the bootstrap test (1,000 bootstrap replications) available in MEGA.
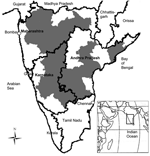
Acute onset of moderate-to-high fever in association with body ache, backache, and headache was recorded. Joint pain of varying severity occurred within 2 days of onset of fever and, in decreasing order of affliction, involved knees, ankles, wrists, hands, and feet. Joint pain was severe and incapacitating and lasted for weeks to months. Inflammation of joints and transient macular rash on earlobes, neck, trunk, and upper extremities were reported for a few patients. Hemorrhage did not occur. The cases were reported predominantly from rural areas; distribution was focal. Multiple cases were recorded in families. All ages and both sexes were affected; significantly more cases occurred in persons aged >15 years (299 [89.8%] of 333, p<0.001). Cases were reported from 11 of 23 districts in Andhra Pradesh, 15 of 27 in Karnataka, and 16 of 35 in Maharashtra (Figure 1). Results of serologic testing and virus isolation are shown in Table 2.
State governments of Andhra Pradesh, Karnataka, and Maharashtra have declared outbreaks of CHIKV. By mid-April, the declared numbers of fever cases associated with this outbreak were >25,000 in Andhra Pradesh, >65,000 in Maharashtra, and >36,000 in Karnataka. In absence of active surveillance for this disease, these numbers may be underestimates.
The predominant mosquito species in the affected areas was Ae. aegypti. Ae. albopictus was either absent or present in negligible numbers. The population of Ae. aegypti was reasonably high in most of the localities; adult household indexes and Breteau indexes, respectively, were 10–60 and 13–75 in Andhra Pradesh, 20–70 and 40–200 in Karnataka, and 10–30 and 30–50 in Maharashtra. High density of Ae. aegypti populations in affected areas and 23 isolations or detections of CHIKV from adult mosquitoes indicate that this species is the main vector in India. Earlier outbreaks in India were mainly restricted to large cities; in contrast, the current outbreak is predominantly rural.
Anti-CHIKV IgM was detected in 33.5% to 41.9% of patients tested. The finding of antibodies to dengue virus in 0.9% to 9.9% of patients and to CHIKV and dengue virus in 0.4% to 4.3% of patients indicates that these viruses cocirculate in the area. Nine patients whose acute-phase serum sample was negative had anti-CHIKV IgM in the early convalescent-phase sample, collected during the second week of illness.
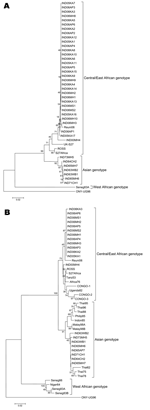
NS4-based phylogenic analysis identified the Yawat isolate (2000) from Maharashtra as central/East African genotype, not Asian genotype as reported earlier (14). This finding led us to resequence all isolates in our repository. Phylogenic analyses based on NS4 (Figure 2A) and E1 regions (Figure 2B) yielded identical results. The Indian viruses isolated from 1963 through 1973 belonged to the Asian genotype, whereas the current isolates from the 3 Indian states and the Yawat isolate belonged to the central/East African genotype. Within the Asian genotype, all older isolates (India 1963–1973 and Thailand 1962–1978) clustered together, whereas later isolates from the Philippines (1985), Indonesia (1985), Thailand (1988, 1995, 1996), and Malaysia (1998) formed a distinct cluster. The sequence from Reunion Islands, which represents a recent outbreak of the disease (GenBank accession no. DQ443544), also grouped with the recent Indian isolates. Percentage nucleotide identity within earlier (1963–1973) and recent (2005–2006) Indian isolates was 99.71% ± 0.16% and 99.94% ± 0.05%, respectively, whereas percentage nucleotide identity between these isolates was 96.11% ± 1.09%. The 2005–2006 Indian isolates were 98.61% ± 0.6% and 98.95% ± 0.57% identical with the Reunion and Yawat isolates, respectively.
This report confirms CHIKV as the causative agent for large outbreaks of fever with arthralgia and arthritis in 3 Indian states. Thus, chikungunya fever has emerged in outbreak form after 32 years.
The current epidemic is caused by central/East African genotype of CHIKV. That the Yawat isolate is grouped with central/East African genotype suggests that this genotype had been introduced >5 years before the current outbreaks. In this context, determining the genotype of currently circulating strains in Southeast Asia and understanding the modes of transportation of this strain in India and the conditions favoring such large outbreaks would be worthwhile.
Mr Yergolkar is in charge of the field station of the National Institute of Virology at Bangalore and is responsible for providing year-round arboviral diagnoses in Karnataka State. He is also in charge of polio surveillance activity as part of the World Health Organization chain of laboratories. His research interest is the epidemiology of arboviruses.
Acknowledgment
We thank the staff of the health departments of Maharashtra, Karnataka, and Andhra Pradesh for help and support during the field investigations. We also thank M.V. Joshi and P.V.M. Mahadev for valuable guidance during field investigations, Kailash B. Gadekar and Ruben P. George for technical support during field investigations, and Santosh M. Jadhav and Atul M. Walimbe for data analysis and illustrations.
References
- Jupp PG, McIntosh BM. Chikungunya virus disease. In: Monath TP, editor. The arboviruses: epidemiology and ecology. Vol. II. Boca Raton (FL): CRC Press; 1988. p. 137–57.
- Mackenzie JS, Chua KB, Daniels PW, Eaton BT, Field HE, Hall RA, Emerging viral diseases of Southeast Asia and the Western Pacific. Emerg Infect Dis. 2001;7:497–504. DOIPubMedGoogle Scholar
- Kit LS. Emerging and re-emerging diseases in Malaysia. Asia Pac J Public Health. 2002;14:6–8. DOIPubMedGoogle Scholar
- Laras K, Sukri NC, Larasati RP, Bangs MJ, Kosim R, Djauzi , Tracking the re-emergence of epidemic chikungunya virus in Indonesia. Trans R Soc Trop Med Hyg. 2005;99:128–41. DOIPubMedGoogle Scholar
- Enserink M. Infectious diseases. Massive outbreak draws fresh attention to little-known virus. Science. 2006;311:1085. DOIPubMedGoogle Scholar
- Nathan MB, Knudsen AB. Aedes aegypti infestation characteristics in several Caribbean countries and implications for integrated community based control. J Am Mosq Control Assoc. 1991;7:400–4.PubMedGoogle Scholar
- Igarashi A. Isolation of a Singh's Aedes albopictus cell clone sensitive to dengue and chikungunya viruses. J Gen Virol. 1978;40:531–44. DOIPubMedGoogle Scholar
- Gadkari DA, Shaikh BH. IgM antibody capture ELISA in the diagnosis of Japanese encephalitis, West Nile and dengue virus infections. Indian J Med Res. 1984;80:613–9.PubMedGoogle Scholar
- Hundekar SL, Thakare JP, Gokhale MD, Barde PV, Argade SV, Mourya DT. Development of monoclonal antibody based antigen capture ELISA to detect chikungunya virus antigen in mosquitoes. Indian J Med Res. 2002;115:144–8.PubMedGoogle Scholar
- Kuberski TT, Rosen L. A simple technique for the detection of dengue antigen in mosquitoes by immunofluorescence. Am J Trop Med Hyg. 1977;26:533–7.PubMedGoogle Scholar
- Sanchez-Seco MP, Rosario D, Quiroz E, Guzman G, Tenorio A. A generic nested-RT-PCR followed by sequencing for detection and identification of members of the Alphavirus genus. J Virol Methods. 2001;95:153–61. DOIPubMedGoogle Scholar
- Hasebe F, Parquet MC, Pandey BD, Mathenge EG, Morita K, Balasubramaniam V, Combined detection and genotyping of Chikungunya virus by a specific reverse transcription-polymerase chain reaction. J Med Virol. 2002;67:370–4. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. DOIPubMedGoogle Scholar
- Yadav P, Shouche YS, Munot HP, Mishra AC, Mourya DT. Genotyping of chikungunya virus isolates from India during 1963–2000 by reverse transcription–polymerase chain reaction. Acta Virol. 2003;47:125–7.PubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 12, Number 10—October 2006
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
A.C. Mishra, 20-A, Dr Ambedkar Rd, National Institute of Virology, Pune, India-411001A.C. Mishra, 20-A, Dr Ambedkar Rd, National Institute of Virology, Pune, India-411001
Top