Volume 12, Number 10—October 2006
Letter
Ciprofloxacin-resistant Salmonella Kentucky in Travelers
Figure
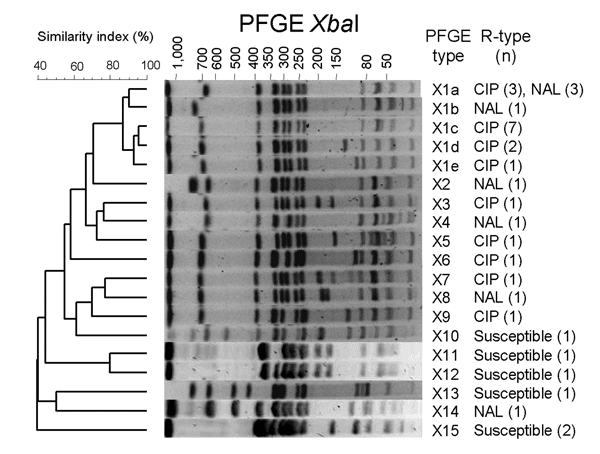
Figure. Dendrogram generated by BioNumerics version 4.1 (Applied Maths, Sint-Martens-Latem, Belgium) showing the results of cluster analysis on the basis of XbaI pulsed-field gel electrophoresis (PFGE) of Salmonella enterica serotype Kentucky isolates. Similarity analysis was performed by using the Dice coefficient, and clustering was performed by the unweighted pair-group method with arithmetic means with an optimization parameter of 0.5% and a 0.5% band position tolerance. The different PFGE profiles, the phenotypes of resistance to quinolones (R-type), and corresponding number of isolates are indicated. CIP, ciprofloxacin; NAL, nalidixic acid.
Page created: November 10, 2011
Page updated: November 10, 2011
Page reviewed: November 10, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.