Volume 13, Number 12—December 2007
Letter
Fatal Streptococcus equi subsp. ruminatorum Infection in a Man
Figure
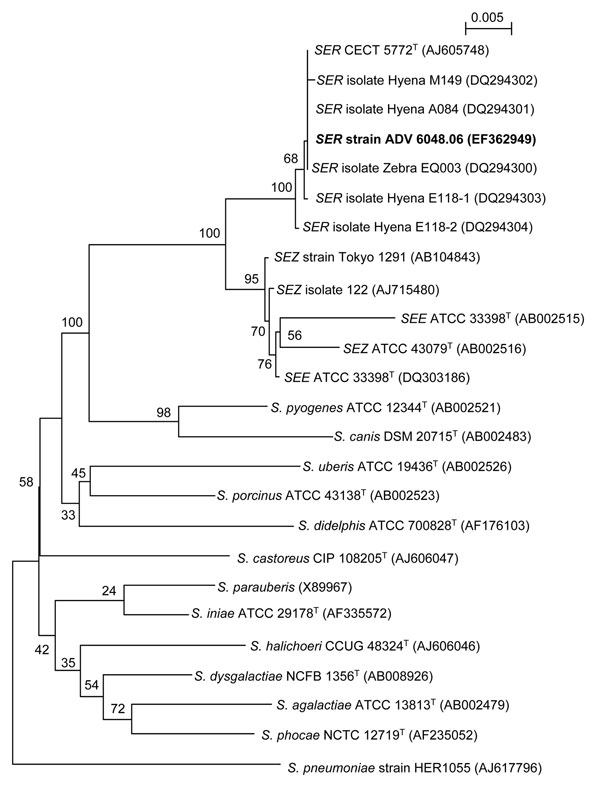
Figure. Neighbor-joining tree showing the phylogenetic placement of strain ADV 6048.06 (boldface) among members of the Streptococcus equi species in the pyogenic group of streptococci. Twenty-three 16S rRNA gene sequences selected from the GenBank database were aligned with that of strain ADV 6048.06 by using ClustalX 1.83 (available from http://bips.u-strasbg.fr/fr/documentation/ClustalX). Alignment of 1,263 bp was used to reconstruct phylogenies by using PHYLIP v3.66 package (http://evolution.genetics.washington.edu/phylip.html). The neighbor-joining tree was constructed with a distance matrix calculated with F84 model. Numbers given at the nodes are bootstrap values estimated with 100 replicates. S. pneumoniae is used as outgroup organism. Accession numbers are indicated in brackets. The scale bar indicates 0.005 substitutions per nucleotide position. Maximum likelihood and parsimony trees were globally congruent with the distance tree and confirmed the placement of the strain ADV 6048.06 in the S. equi subspecies ruminatorum (SER) lineage. SEZ, S. equi subspecies zooepidemicus.