Volume 13, Number 3—March 2007
Dispatch
Cocirculation of 2 Genotypes of Toscana Virus, Southeastern France
Cite This Article
Citation for Media
Abstract
Toscana virus (TOSV), an arthropodborne phlebovirus transmitted by sandflies, can cause febrile illness and meningitis. The vector of TOSV in France was unknown. We detected TOSV RNA in 2 (female Phlebotomus perniciosus) of 61 pools of sandflies captured in southeastern France. Two genotypes of TOSV were identified.
Toscana virus (TOSV) is an arthropodborne virus first isolated in 1971 in central Italy from the sandfly Phlebotomus perniciosus, then from Ph. perfiliewi (1,2). In Spain, TOSV has been isolated from phlebotomine flies, but species identification was not performed (3). TOSV has not been detected in or isolated from sandflies in France.
Most clinical and epidemiologic studies on TOSV have been conducted in Italy, although data from other Mediterranean countries have recently been published (4). TOSV has a tropism for the central nervous system and is a major cause of meningitis and encephalitis in the countries in which the virus circulates. Cases of TOSV infection acquired in the south of France indicate that TOSV circulates in the region (5,6). The aim of our study was to identify the presence of TOSV in Phlebotomus spp. collected in southeastern France and to investigate French TOSV at the genetic level. For that purpose, we compared viral sequences from sandflies and from French patients with other sequence data previously reported from Italy and Spain.
Sandflies were trapped during 4 days in July 2005 in 2 cities in southern France (Marseille and Nice) by using techniques previously reported (7) (Figure 1). Adult flies shelter during the day in dark, quiet, and humid places, and eggs are laid in terrestrial microhabitats rich in organic matter that provides food for larvae, including soils containing herbivorous animal feces. Therefore, traps were placed at dawn in or near animal housing facilities (horse stalls, rabbit hutches, hen houses) or in areas where visceral canine leishmaniasis is endemic (8). Each morning, sandflies were collected and identified morphologically, by dissecting genital organs, according to morphologic taxonomic keys (9). Sandflies were pooled (up to 30 individual flies) according to trapping origin, species, and gender (Table) in 1.5-mL tubes and processed as described (7).
A variety of primers targeting different genes were used: primers specific for TOSV in the polymerase gene (10) and primers designed in this study from the alignment of nucleoprotein sequences of selected phleboviruses (retrieved from GenBank) specific for the species Sandfly fever Naples virus (SFNV-S1 [5′-CTTYTTRTCYTCYCTRGTGAAGAA-3′], SFNV-R1 [5′-ATGATGAAGAARATGTCAGAGAA-3′], SFNV-S2 [5′-GCRGCCATRTTKGGYTTTTCAAA-3′], SFNV-R2 [5′-CCTGGCAGRGACACYATCAC-3′]). The reverse-transcription–PCR (RT-PCR) was performed by using the Access RT-PCR kit (Promega, Madison, WI, USA) according to the manufacturer’s recommendations. L and S RNA primers were used at 0.8 μmol and 0.4 μmol per reaction, respectively. The cycling program of the RT-PCR reaction consisted of 48°C for 45 min and 94°C for 2 min, followed by 40 cycles at 94°C for 30 s, the annealing temperature for 1 min, and 68°C for 45 s, with a final elongation step at 68°C for 7 min.
Using ground material obtained from pools AK and AR and patients’ samples, we injected Vero-E6 cells (protocol available on request). Most sandflies belonged to the Ph. perniciosus species (Table, Figure 1), 483 (96.8%) in Marseille, 110 (62.5%) in Nice. Two other species of phlebotomine flies were identified, Ph. mascitii (n = 4, 0.8%), and Ph. ariasi (n = 3, 0.6%) in Marseille. In addition, 123 Sergentomyia minuta were identified and led to the detection of TOSV RNA, as recently reported (7). Finally, 61 pools of Phlebotomus spp. were organized to be tested for the presence of TOSV RNA.
We detected TOSV RNA by both PCRs in 2 pools (AK and AR) of female Ph. perniciosus trapped in the neighborhood of Marseille. AK and AR sequences were distinct from each other (1.4% and 1% nucleotide divergence for S and L RNA sequences, respectively), from other sequences available in GenBank, and from those determined during this study or in previous studies (0.7%–21.5% and 17.4% nucleotide divergence for S and L RNA sequences, respectively) (7).
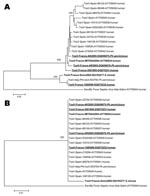
Sequences from 3 TOSV-infected patients who lived in the region of Marseille were included in the genetic analysis. For 2 patients, sequences were obtained after virus isolation in Vero cells: strain IMTSSA/2004 (GenBank accession no. AY766034) (6) and strain 1500590 (GenBank accession no DQ975232–975233, this study). Another sequence was determined from direct amplification of a cerebrospinal fluid sample (GenBank accession no. DQ975231, this study). Nucleotide distances (Appendix Figure) clearly allowed discrimination of the 2 genotypes previously described as the Italian and the Spanish genotypes by reference to the origin of viruses. Sequences obtained from sandfly and clinical material collected in France indicated that both genotypes are circulating in southern France. Indeed, Ph. perniciosus specimens AK and AR and human samples 5501805 and IMTSSA/2004 displayed high identity levels with TOSV of Spanish origin. In contrast, S. minuta specimen Smin2005 and human sample 1500590 were more closely related to the Italian strain Iss.Phl3 (Appendix Figure). Phylogenetic analysis of nucleotide sequences (Figure 2A) showed that Spanish, French, and Italian TOSV and SFNV formed a cluster that included members of the species Sandfly fever Naples virus, for which genetic data were available. Such analysis confirmed that the 2 French TOSV sequences detected in Ph. perniciosus (AK and AR) and the 2 TOSV sequences from clinical samples (IMTSSA2004 and 5501805) grouped with Spanish TOSV (100% bootstrap support), while S. minuta specimen Smin2005 and human sample 1500590 clustered with Italian TOSV sequences. Topology analysis indicated that the 4 French TOSV most closely related to Spanish TOSV root the Spanish sequences (Figure 2A). Together, all these sequences displayed a cline topology. Further studies, with more sequences, are needed to investigate this finding. As previously reported (3), despite significant variability between TOSV isolates at the nucleotide level (up to 13.2%), phylogenetic analysis of amino acid sequences did not allow discrimination between the Italian and Spanish genotypes (Appendix Figure, Figure 2B). Indeed, in the nucleoprotein region considered for analysis, all TOSV amino acid sequences were 100% identical, except that obtained from S. minuta, which diverged by 1 amino acid (data not shown).
This study provided genetic evidence, from the analysis of 2 independent genes, that TOSV circulates in populations of Ph. perniciosus in southeastern France. The absence of detection of TOSV in the Nice region may be due to the small number of sandflies collected. A similar study aimed at collecting a larger number of sandflies is therefore necessary to address whether TOSV circulates in the region of Nice and whether, if it does, it belongs to the Spanish genotype, the Italian genotype, or another yet unrecognized genotype.
The rate of infection observed with TOSV in sandflies collected during this study (2/675) is of the same magnitude of that reported in Spain (3) and lower than rates reported in Italy (2). The distribution and respective frequency of sandfly species in France (this study) and Spain (3) were different: Ph. papatasi (6.8%) and Ph. sergenti (8.5%) were cataloged in Spain, not in France, whereas Ph. mascitii (5.9%) was identified in France, not in Spain. Ph. perfiliewi, known to replicate TOSV in nature in Italy, has not been identified in either French or Spanish studies.
In conclusion, we have provided the first evidence for the circulation of TOSV in Ph. perniciosus in the region of Marseille, southeastern France. We also determined that TOSV belonging to the 2 genotypes previously recognized in Italy and Spain circulates and causes human infections in southeastern France.
Dr Charrel is a virologist who works in a hospital diagnostic laboratory and with a university research group. His research interests are arthropodborne and rodentborne viruses that cause disease in humans, with a special interest in emerging and reemerging viruses such as arenaviruses, flaviviruses, and phleboviruses.
Acknowledgment
This work has been supported in part by the European Commission through RiVigene project, and by the Collection Virale de Biodéfense – Diagnostics.project.
References
- Nichol ST, Beaty BJ, Elliott RM, Goldbach R, Plyusnin A, Schmaljohn CS, Genus Phlebovirus. In Fauquet CM, Mayo MH, Maniloff J, Desselberger U, Ball LA. Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee of the Taxonomy of Viruses; 2005. San Diego: Elsevier Academic Press;2005. pp.709–16.
- Verani P, Ciufolini MG, Caciolli S, Renzi A, Nicoletti L, Sabatinelli G, Ecology of viruses isolated from sand flies in Italy and characterized of a new Phlebovirus (Arabia virus). Am J Trop Med Hyg. 1988;38:433–9.PubMedGoogle Scholar
- Sanbonmatsu-Gamez S, Perez-Ruiz M, Collao X, Sanchez-Seco MP, Morillas-Marquez F, de la Rosa-Fraile M, TOSV in Spain. Emerg Infect Dis. 2005;11:1701–7.PubMedGoogle Scholar
- Charrel RN, Gallian P, Navarro-Mari J-M, Nicoletti L, Papa A, Sánchez-Seco MP, Emergence of Toscana virus in Europe. Emerg Infect Dis. 2005;1:1657–63.
- Hemmersbach-Miller M, Parola P, Charrel RN, Paul Durand J, Brouqui P. Sandfly fever due to TOSV: an emerging infection in southern France. Eur J Intern Med. 2004;15:316–7. DOIPubMedGoogle Scholar
- Peyrefitte CN, Devetakov I, Pastorino B, Villeneuve L, Bessaud M, Stolidi P, TOSV and acute meningitis, France. Emerg Infect Dis. 2005;11:778–80.PubMedGoogle Scholar
- Charrel RN, Izri A, Temmam S, de Lamballerie X, Parola P. Toscana virus RNA in Sergentomyia minuta files. Emerg Infect Dis. 2006;12:1299–300.PubMedGoogle Scholar
- Marty P, Le Fichoux Y, Pratlong F, Gari-Toussaint M. Human visceral leishmaniasis in Alpes-Maritimes, France: epidemiological characteristics for the period 1985–1992. Trans R Soc Trop Med Hyg. 1994;88:33–4. DOIPubMedGoogle Scholar
- Léger N, Pesson B, Madullo-Leblond G, Abonnenc E. Sur la différentiation des femelles du genre Laroussius Nitzelescu, 1931 (Diptera, Phlebotomidae) de la région méditerranéenne. Ann Parasitol Hum Comp. 1983;58:611–23.PubMedGoogle Scholar
- Sanchez Seco MP, Echevarría JM, Hernández L, Estévez D, Navarro Marí JM, Tenorio A. Detection and identification of Toscana and other phleboviruses by RT-PCR assays with degenerated primers. J Med Virol. 2003;71:140–9. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. DOIPubMedGoogle Scholar
Figures
Table
Cite This Article1Equally participated in this work.
2Equally participated in the design and supervision of this study.
Table of Contents – Volume 13, Number 3—March 2007
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Remi N. Charrel, Unité des Virus Emergents, Faculté de Médecine, 27 blvd Jean Moulin, Marseille 13005, France;
Top