Volume 13, Number 3—March 2007
Letter
Novel Hantavirus Sequences in Shrew, Guinea
Figure
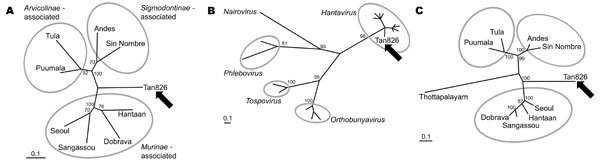
Figure. Maximum likelihood phylogenetic analysis of hantaviruses showing the phylogenetic placement of Tan826 (Tanganya virus, indicated by arrow) based on partial L segment nucleotide (A) and amino acid (B) sequences and partial S segment amino acid sequences (C); GenBank accession nos. EF050454 and EF050455, respectively. The values near the branches represent PUZZLE support values (4) calculated from 10,000 puzzling steps; only values ≥70% are shown. The scale bar indicates an evolutionary distance of 0.1 substitutions per position in the sequence. Gray ellipsoids indicate the 3 major hantavirus groups (panels A and C) or different genera of the Bunyaviridae family (panel B). We used 412 nt (137 aa) of the L segment (nucleotide position 2956–3367, amino acid position 974–1110) and 147 aa (nucleotide position 685–1111, amino acid position 217–364) of the putative nucleoprotein. Fragment positions were defined according to complete sequences of Hantaan virus strain 76-118 (GenBank accession nos. NC_005222 and NC_005218, respectively). The L segment fragment was amplified as described previously (4); for the S segment fragment, highly degenerated primers Cro2F (5′-AGYCCIGTIATGRGWGTIRTYGG-3′) and Cro2R (5′-AIGAYTGRTARAAIGAIGAYTTYTT-3′) were used. TREE-PUZZLE (www.tree-puzzle.de; [4]) was used to calculate the trees with HKY (panel A) and JTT (panels B and C) evolutionary models. Missing parameters were reconstructed from the data set. The following sequences were obtained from GenBank: X55901, AJ410617, X56492, DQ268652, M63194, AJ005637, L37901, AF291704, X14383, U12396, AF484424, AF133128, D10066, X56464, D10759, U15018 for L segment and M14626, L41916, S47716, DQ268650, M32750, Z69991, L25784, AF291702, AY526097 for S segment analysis.
References
- Schmaljohn CS, Hjelle B. Hantaviruses: a global disease problem.Emerg Infect Dis. 1997;3:95–104.PubMedGoogle Scholar
- Kruger DH, Ulrich R, Lundkvist A. Hantavirus infections and their prevention.Microbes Infect. 2001;3:1129–44. DOIPubMedGoogle Scholar
- Mertz GJ, Hjelle BL, Bryan RT. Hantavirus infection.Adv Intern Med. 1997;42:369–421.PubMedGoogle Scholar
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing.Bioinformatics. 2002;18:502–4. DOIPubMedGoogle Scholar
- Klempa B, Fichet-Calvet E, Lecompte E, Auste B, Aniskin V, Meisel H, Hantavirus in African wood mouse, Guinea.Emerg Infect Dis. 2006;12:838–40.PubMedGoogle Scholar
- Carey DE, Reuben R, Panicker KN, Shope RE, Myers RM. Thottapalayam virus: a presumptive arbovirus isolated from a shrew in India.Indian J Med Res. 1971;59:1758–60.PubMedGoogle Scholar
- Zeller HG, Karabatsos N, Calisher CH, Digoutte JP, Cropp CB, Murphy FA, Electron microscopic and antigenic studies of uncharacterized viruses. II. Evidence suggesting the placement of viruses in the family Bunyaviridae.Arch Virol. 1989;108:211–27. DOIPubMedGoogle Scholar
- Xiao SY, Leduc JW, Chu YK, Schmaljohn CS. Phylogenetic analyses of virus isolates in the genus Hantavirus, family Bunyaviridae.Virology. 1994;198:205–17. DOIPubMedGoogle Scholar
- Hutterer R. Order Soricomorpha. In: Wilson DE, Reeder DM, editors. Mammal species of the world: A taxonomic and geographic reference, 3rd ed, vol.1. Baltimore: Johns Hopkins University Press; 2005. p. 220–311.
- Chu YK, Jennings G, Schmaljohn A, Elgh F, Hjelle B, Lee HW, Cross-neutralization of hantaviruses with immune sera from experimentally infected animals and from hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome patients.J Infect Dis. 1995;172:1581–4.PubMedGoogle Scholar