Volume 14, Number 11—November 2008
Dispatch
Porcine Respiratory and Reproductive Syndrome Virus Variants, Vietnam and China, 2007
Cite This Article
Citation for Media
Abstract
We characterized isolates from porcine respiratory and reproductive syndrome virus epidemics in Vietnam and China in 2007. These isolates showed ≈99% identity at the genomic level. Genetic analysis indicated that they share a discontinuous deletion of 30 aa in nonstructural protein 2, which indicates that identical variants emerged in Vietnam and China.
Porcine respiratory and reproductive syndrome (PRRS) is one of the most economically influential infectious diseases in the industry of swine cultivation. The etiologic agent is PRRS virus (PRRSV), a member of the family Arteriviridae in the order Nidovirales (1,2). Genomic analysis of PRRSVs has shown that the virus genome varies from 15 kb to 15.5 kb and comprises at least 8 open reading frames (ORFs) that encode nearly 20 mature proteins (3). PRRSVs with different geographic origins can be classified into 2 major genotypes, the European type (type I, EU-type) (4) and North American type (type II, NA-type) (5). Nonstructural protein 2 (Nsp2) (6) and glycoprotein 5 (encoded by ORF5) (7), are regarded as 2 regions of high heterogeneity that are involved in the pathogenicity of PRRSV strains.
In general, PRRSV has caused either respiratory failure in neonates or abortions in sows during sporadic PRRS outbreaks worldwide (2). The unprecedented large-scale PRRS outbreaks in 2006 swept over nearly half of the People’s Republic of China and involved >2,000,000 pigs, which posed great concern to the global swine industry and to public health (8). Subsequent genomic analysis showed that all the PRRSVs isolated from this outbreak (8) share a unique discontinuous deletion of 30 aa in Nsp2. In previous studies, only 2 virulent NA-type PRRSV strains (P129 and MN184) had been suggested to carry deletions in the Nsp2 protein (different from our 30-aa deletion), a highly variable region that contributes to the virulence and PRRSV genotyping (9,10).
A suspected PRRS outbreak was observed in Vietnam in 2007, and further spread of the disease has been found in China in 2007. To elucidate the characteristics of these outbreaks and the PRRSVs isolated, we conducted a detailed investigation.
In March 2007, the suspected PRRS-like disease was initially found in Hai Duong Province, the northern province of Vietnam. The disease later spread to nearly the entire country and affected at least 65,000 pigs. Meanwhile, the recurrence of PRRSV infections in China was officially announced in May 2007 by the Chinese Ministry of Agriculture.
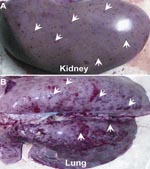
The affected pigs exhibit the clinical features of high fever, depression, and shivering. Viscera (e.g., brain, kidney, lung, heart, liver, and spleen) were sampled from those dead pigs from different provinces in Vietnam and China. Pathologic examination showed severe lesions viscera, for example, blood spots in the kidney and hemorrhages in the lung (Figure 1, panels A, B), findings similar to those observed in the 2006 PRRS outbreaks in China (8,11). The inocula (from each representative specimen) were propagated in Marc-145 cells (QIAGEN, Hilden, Germany) for the viral genomic RNA isolation. Based on our previous observation of the 2006 PRRS epidemics in China, we designed 2 pairs of specific primers (Nsp2-F: 5′-AAA GAC CAG ATG GAG GAG GA-3′ and Nsp2-R: 5′-GAG CTG AGT ATT TTG GGC GTG-3′; orf5-F: 5′-ATG TTG GGG AAG TGC TTG ACC-3′ and orf5-R: 5′-CTA GAG ACG ACC CCA TTG TTC CGC-3′). They correspond to the DNA fragment covering a putative discontinuous deletion of 30 aa in Nsp2 (666 bp), and ORF5 in full length, respectively. PCR-based detection showed that all the samples collected so far, are positive for PRRSV, but negative for African swine fever, classical swine fever, and foot-and-mouth disease viruses.
To test the virulence of Vietnam PRRSV isolates, 6 specific pathogen–free piglets were challenged with a representative strain (termed 07QN, which had been isolated from a dead pig in Quang Nam Province, Vietnam). The piglets were monitored for clinical signs every 6 hours. All procedures were conducted in a facility qualified with Biosafety Level 3 and approved by Vietnamese Committee for Approval of Drugs and Cosmetics ethics committee. As we expected, the piglets reproduced the serious symptoms similar to those observed in our field investigation, which indicated that the viral agent is highly pathogenic.
To better understand the genetic relationship of Vietnamese PRRSV isolates to the 2006 and 2007 Chinese isolates, 5 strains of PRRSVs (1 isolate from Vietnam, 07QN, and 4 newly isolated isolates from China, 07HEBTJ, 07BJ, 07HEN, and 07NM) were subjected to whole genome sequencing as described (8). The viral genomes were found to be >15,300 bp (GenBank accession nos. are available from the authors). Bioinformatics analysis further revealed that the 2007 Vietnamese isolate (07QN) and 2007 viruses (e.g., 07BJ) have 99% identity to 2006 PRRS isolates in China at the level of nucleic acid sequences. Whole genome–based phylogenetic relationship also showed that all these viral isolates are grouped into the same subclade in the type II genotype (Figure 2). All of the 2007 strains, together with those Chinese viruses collected in 2006, were found to be similar to 3 Chinese strains reported previously (HB-1, HB-2, and CH-1a) (3,12) and 1 NA-type virulent strain, P129 (8,9).
In addition, >30 sequences of orf5, a highly variant gene, were sequenced, and ORF5-based genotyping also supported the classification of these 2007 isolates (from Vietnam and China) into a subgroup of type II, while far from other subclades with VR-2332, a prototype of type II (not shown). Moreover, the multiple alignments of Nsp2 demonstrated that the 2007 PRRSV isolates from Vietnam (51 total), together with 2007 Chinese strains (6 total) are nearly identical and share a discontinuous deletion of 30 aa at the position of 482 aa and 534–562 aa, which is consistent with deletions in the strains from the 2006 epidemic in China (8).
PRRS has become a serious challenge to the global pig industry, causing serious economic losses (2). In particular, the unparalleled 2006 PRRS outbreaks engulfed China, and adult pigs were not exempt, which indicates that these variants may have evolved to exhibit new characteristics of pathogenicity (8). These outbreaks caused extensive concern worldwide (13). Even though the deletion of 30 aa in Nsp2 has been proposed as a potential virulence factor (8), the molecular mechanisms underlying its high virulence are yet to be elucidated.
Origin of these lethal variants in Vietnam is still obscure, although Kamakawa et al. (14) suggested that PRRSV may have been present in Vietnam before 1999. Convergent, but separate, evolution of PRRSVs in Vietnam and China may explain in part the emergence of the nearly identical PRRSV variants in these 2 neighboring countries, although this hypothesis does not rule out the possibility that 2006 Chinese PRRSV variants were transmitted into Vietnam and then circulated rapidly. Additionally, intraprovincial and interprovincial transportation of live pigs and the similar climate in Vietnam and China may have contributed to these outbreaks (15).
In summary, our findings provided robust evidence that nearly identical variants of NA-type PRRSVs are the causative pathogens that triggered PRRS epidemics in Vietnam and China in 2007. This finding highlights the importance of prevention and control of this highly transmissible infectious agent.
Dr Feng is a microbiologist at Chinese Academy of Sciences in Beijing, focusing on emerging and reemerging infectious diseases. His research interests include PRRSV, Streptococcus suis, and avian influenza.
Acknowledgment
This work was supported by National Key Technologies R&D Programs (2007BAD86B04) and National Basic Research Program (Project 973) of China (grant no. 2005CB523001). G.F.G. is a distinguished young investigator of the National Natural Science Foundation of China (grant no. 30525010).
References
- Zimmerman JJ, Yoon KJ, Wills RW, Swenson SL. General overview of PRRSV: a perspective from the United States. Vet Microbiol. 1997;55:187–96. DOIPubMedGoogle Scholar
- Gao ZQ, Guo X, Yang HC. Genomic characterization of two Chinese isolates of porcine respiratory and reproductive syndrome virus. Arch Virol. 2004;149:1341–51. DOIPubMedGoogle Scholar
- Wensvoort G. Lelystad virus and the porcine epidemic abortion and respiratory syndrome. Vet Res. 1993;24:117–24.PubMedGoogle Scholar
- Nelson EA, Christopher-Hennings J, Drew T, Wensvoort G, Collins JE, Benfield DA. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J Clin Microbiol. 1993;31:3184–9.PubMedGoogle Scholar
- Fang Y, Kim DY, Ropp S, Steen P, Christopher-Hennings J, Nelson EA, Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 2004;100:229–35. DOIPubMedGoogle Scholar
- An TQ, Zhou YJ, Liu GQ, Tian ZJ, Li J, Qiu HJ, Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: coexistence of two NA-subgenotypes with great diversity. Vet Microbiol. 2007;123:43–52. DOIPubMedGoogle Scholar
- Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007;2:e526. DOIPubMedGoogle Scholar
- Yoo D, Welch SK, Lee C, Calvert JG. Infectious cDNA clones of porcine reproductive and respiratory syndrome virus and their potential as vaccine vectors. Vet Immunol Immunopathol. 2004;102:143–54. DOIPubMedGoogle Scholar
- Han J, Wang Y, Faaberg KS. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 2006;122:175–82. DOIPubMedGoogle Scholar
- Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis. 2007;13:1434–6.PubMedGoogle Scholar
- Chen J, Liu T, Zhu CG, Jin YF, Zhang YZ. Genetic variation of Chinese PRRSV strains based on ORF5 sequence. Biochem Genet. 2006;44:425–35.PubMedGoogle Scholar
- Normile D. Virology. China, Vietnam grapple with ‘rapidly evolving’ pig virus. Science. 2007;317:1017. DOIPubMedGoogle Scholar
- Kamakawa A, Ho TV, Yamada S. Epidemiological survey of viral diseases of pigs in the Mekong delta of Vietnam between 1999 and 2003. Vet Microbiol. 2006;118:47–56. DOIPubMedGoogle Scholar
- Hermann J, Hoff S, Muñoz-Zanzi C, Yoon KJ, Roof M, Burkhardt A, Effect of temperature and relative humidity on the stability of infectious porcine reproductive and respiratory syndrome virus in aerosols. Vet Res. 2007;38:81–93. DOIPubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 14, Number 11—November 2008
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Kegong Tian, China Animal Disease Control Center, Beijing 100094, People’s Republic of China;
Top