Volume 14, Number 5—May 2008
Letter
Hantavirus Outbreak, Germany, 2007
Figure
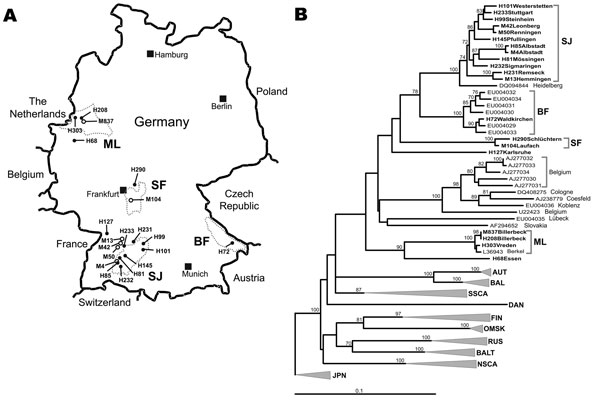
Figure. A) Map of Germany showing origins of viral sequences from the 2007 outbreak. H, sequences of human origin; M, sequences of rodent origin (Myodes glareolus). Dotted circles mark the outbreak regions characterized by particular virus sequence clusters; SJ, Swabian Jura; BF, Bavarian Forest; SF, Spessart Forest; ML, Munsterland. B) Neighbor-joining phylogenetic tree (TN93 evolutionary model) of European Puumala virus (PUUV) strains based on partial sequences of the S segment (557 nt, position 385–941). Bootstrap values >70%, calculated from 10,000 replicates, are shown at the tree branches. PUUV-like sequences from Japan (JPN) were used as an outgroup. Sequences taken from GenBank are indicated by their accession numbers. New sequences from this study are given in boldface. Accession numbers of new sequences are H101, EU266757; H233, EU266758; H99, EU266759; M42, EU085563; M50, EU085565 ; H145, EU266760; H85, EU266761; M4, EU266762; H81, EU266763; H232, EU266764; H231, EU266765; M13, EU085558; H72, EU266766; H290, EU266767; M104, EU246963; H127, EU266768; M837, EU266769; H208, EU266770; H303, EU266771; H68, EU266772. For clarity, previously characterized PUUV clades from other parts of Europe are shown in simplified form. However, the complete dataset of PUUV sequences as presented by Schilling et al. (6) was used to calculate the tree. Previously defined lineages are indicated by abbreviated names: AUT, Austrian; BAL, Balkan; BALT, Baltic; DAN, Danish; FIN, Finnish; NSCA, North Scandinavian; OMSK, Russian from Omsk region; RUS, Russian; SSCA, South Scandinavian. Scale bar indicates an evolutionary distance of 0.1 substitutions per position.
References
- Kruger DH, Ulrich R, Lundkvist A. Hantavirus infections and their prevention. Microbes Infect. 2001;3:1129–44. DOIPubMedGoogle Scholar
- Robert Koch Institute. Berlin, Germany. Epidemiologisches Bulletin. 2008;(13) [cited 2008 <ar 28]. Available from http://www.rki.de
- Abu Sin M, Stark K, van Treek U, Dieckmann H, Uphoff H, Hautmann W, Risk factors for hantavirus infection in Germany, 2005. Emerg Infect Dis. 2007;13:1364–6.PubMedGoogle Scholar
- Meisel H, Wolbert A, Razanskiene A, Marg A, Kazaks A, Sasnauskas K, Development of novel immunoglobulin G (IgG), IgA, and IgM enzyme immunoassays based on recombinant Puumala and Dobrava hantavirus nucleocapsid proteins. [PMID 17021245]. Clin Vaccine Immunol. 2006;13:1349–57. DOIPubMedGoogle Scholar
- Kramski M, Meisel H, Klempa B, Kruger DH, Pauli G, Nitsche A. Detection and typing of human pathogenic hantaviruses by real-time reverse transcription-PCR and pyrosequencing. Clin Chem. 2007;53:1899–905. DOIPubMedGoogle Scholar
- Schilling S, Emmerich P, Klempa B, Auste B, Schnaith E, Schmitz H, Hantavirus disease outbreak in Germany: limitations of routine serological diagnostics and clustering of virus sequences of human and rodent origin. J Clin Microbiol. 2007;45:3008–14. DOIPubMedGoogle Scholar
- Pilaski J, Feldmann H, Morzunov S, Rollin PE, Ruo SL, Lauer B, Genetic identification of a new Puumala virus strain causing severe hemorrhagic fever with renal syndrome in Germany. J Infect Dis. 1994;170:1456–62.PubMedGoogle Scholar
- Bahr U, Zeier M, Muranyi W. Characterization of a new Puumala virus genotype associated with hemorrhagic fever with renal syndrome. Virus Genes. 2006;33:229–34. DOIPubMedGoogle Scholar
- Zoller L, Faulde M, Meisel H, Ruh B, Kimmig P, Schelling U, Seroprevalence of hantavirus antibodies in Germany as determined by a novel recombinant enzyme immunoassay. Eur J Clin Microbiol Infect Dis. 1995;14:305–13. DOIPubMedGoogle Scholar
- Essbauer S, Schmidt J, Conraths FJ, Friedrich R, Koch J, Hautmann W, A new Puumala hantavirus subtype in rodents associated with an outbreak of Nephropathia epidemica in South-East Germany in 2004. Epidemiol Infect. 2006;134:1333–44. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.
2Current affiliation: Bernhard-Nocht-Institute for Tropical Medicine, Hamburg, Germany.