Volume 15, Number 12—December 2009
Dispatch
Sympatry of 2 Hantavirus Strains, Paraguay, 2003–2007
Figure 2
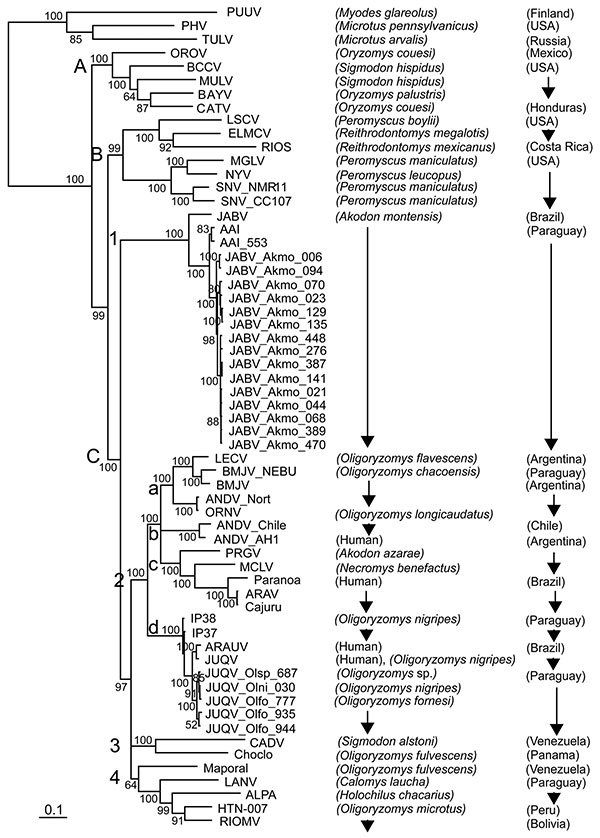
Figure 2. Phylogenetic tree based on Bayesian analysis of the small (S) segment of American hantaviruses, Paraguay, 2003–2007. The tree is based on 1,014 nucleotides of partial S segment of North and South American hantaviruses. Clades (upper case letters), subclades (numbers), and groups (lower case letters) are indicated on the left. Numerical values at the nodes indicate posterior probabilities that supported each interior branch. Scale bar indicates mean number of nucleotide substitutions per site. Sample identifier numbers are the same as in Figure 1. Alignment and editing of nucleotide sequences were conducted by using Vector NTI version 10.3.1 (Invitrogen, Carlsbad, CA, USA). Puumala virus (PUUV) was used as an outgroup for phylogenetic analyses. Estimation of the suitable model of nucleotide substitution and phylogenetic analysis were conducted by using Modeltest version 3.6 (http://darwin.uvigo.es/software/modeltest.html), PAUP* 4.0b10 (http://paup.csit.fsu.edu), and MrBayes version 3.1.2 (http://mrbayes.csit.fsu.edu). A Bayesian analysis was conducted under the general time reversible + invgamma model. Two runs of 4 chains each (1 cold, 3 heated, temperature 0.20) were run for 1 million generations, and trees were sampled every 100 generations. Convergence was assessed by using the average standard deviation in partition frequency values across independent analyses with a threshold value of 0.01; burn-in was set to 25%. Nucleotide sequences of specimens collected at Reserva Natural del Bosque Mbaracayú, Departamento Canindeyú, are indicated by acronyms of related viruses, rodent species, and collection numbers. Hantavirus strain abbreviations and GenBank accession nos. are AAI (Ape Aime) virus from Paraguay [DQ345764]; ALPA (Alto Paraguay) virus from Paraguay [DQ345762]; ANDV_AH1 (Andes virus, strain AH-1) from Argentina [AF324902]; ANDV_Chile (Andes virus, strain Chile 9717869) [AF291702]; ANDV_Nort (Andes virus, strain Nort) from Argentina [AF325966]; ARAV (Araraquara virus) from Brazil [AF307325]; ARAUV (Araucaria virus) from Brazil [AY740633]; BAYV (Bayou virus) from the United StatesA [L36929]; BCCV (Black Creek Canal virus) from the United States [L39949]; BMJV (Bermejo virus) from Argentina [AF482713]; BMJV-ÑEBU (Bermejo virus, strain Ñeembucú) from Paraguay [DQ345763]; Cajuru (Araraquara-like virus, strain Cajuru) from Brazil [EF571895]; CATV (Catacamas virus) from Honduras [DQ256126]; CADV (Cano Delgadito virus) from Venezuela [DQ285566]; Choclo virus from Panama [DQ285046]; ELMCV (El Moro Canyon virus) from the United States [U11427]; HTN-007 virus from Peru [AF133254]; IP37 (Itapúa virus, strain 37) from Paraguay [DQ345765]; IP38 (Itapúa virus, strain 38) from Paraguay [DQ345766]; Jaborá (JAB) virus from Brazil [EF492471]; JUQV (Juquitiba virus) from Brazil [EF492472]; LECV (Lechiguanas virus) from Argentina [AF482714]; MULV (Muleshoe virus) from the United States [U54575]; LANV (Laguna Negra virus) from Paraguay [AF005727]; LSCV (Limestone Canyon virus) from the United States [AF307322]; MCLV (Maciel virus) from Argentina [AF482716]; Maporal virus from Venezuela [AY267347]; MGLV (Monongahela virus) from the United States [U32591]; NYV (New York virus) from the United States [U47135]; OROV (Playa de Oro virus) from the Mexico [EF534077–79]; ORNV (Oran virus) from Argentina [AF482715]; Paranoa virus from Brazil [EF576661]; PHV (Prospect Hill virus) from the United States [U47136]; PRGV (Pergamino virus) from Argentina [AF482717]; PUUV from Finland [X61035]; RIOMV (Rio Mamore virus) from Bolivia [U52136]; RIOS (Rio Segundo) virus from Costa Rica [GenBank U18100 (S segment)]; SNV_CC107 (Sin Nombre virus, strain CC107) from the United States [L33683]; SNV_NMR11 (Sin Nombre virus, strain NMR11) from the United States [L37904]; and TULV (Tula virus) from Russia [Z30941]. OLFO, Oligoryzomys fornesi; OLNI, O. nigripes; OLSP, Oligoryzomys sp.; AKMO, Akodon montensis).