Volume 15, Number 12—December 2009
Dispatch
Mopeia Virus–related Arenavirus in Natal Multimammate Mice, Morogoro, Tanzania
Figure 2
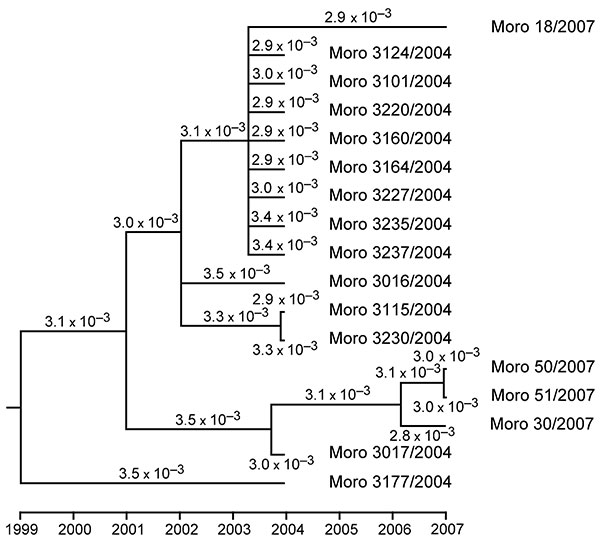
Figure 2. Phylogenetic tree and molecular clock of Morogoro virus based on partial large gene sequences of 17 strains (340 nucleotides; GenBank accession nos. EU914104 and EU914107–EU914122). Phylogeny was inferred with the BEAST v1.4.8 package (11) under assumption of a relaxed lognormal molecular clock and general time reversible substitution model with gamma-distributed substitution rate variation among sites. Branches with posterior probability <0.5 were collapsed. The substitution rate per site and year is indicated for each branch. Node ages and rates are median values. Variation in rates among branches is low as calculated with Tracer program (beast.bio.ed.ac.uk/Tracer) indicating a molecular clock in the evolution of Morogoro virus. The same tree topology with similar substitution rates was obtained when assuming the Hasegawa-Kishino-Yano substitution model (not shown).
References
- Delgado S, Erickson BR, Agudo R, Blair PJ, Vallejo E, Albarino CG, Chapare virus, a newly discovered arenavirus isolated from a fatal hemorrhagic fever case in Bolivia. PLoS Pathog. 2008;4:e1000047. DOIPubMedGoogle Scholar
- Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Genetic detection and characterization of Lujo virus, a new hemorrhagic fever–associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. DOIPubMedGoogle Scholar
- Wulff H, McIntosh BM, Hamner DB, Johnson KM. Isolation of an arenavirus closely related to Lassa virus from Mastomys natalensis in south-east Africa. Bull World Health Organ. 1977;55:441–4.PubMedGoogle Scholar
- Johnson KM, Taylor P, Elliott LH, Tomori O. Recovery of a Lassa-related arenavirus in Zimbabwe. Am J Trop Med Hyg. 1981;30:1291–3.PubMedGoogle Scholar
- Gonzalez JP, McCormick JB, Saluzzo JF, Herve JP, Georges AJ, Johnson KM. An arenavirus isolated from wild-caught rodents (Pramys species) in the Central African Republic. Intervirology. 1983;19:105–12. DOIPubMedGoogle Scholar
- Lecompte E, ter Meulen J, Emonet S, Daffis S, Charrel RN. Genetic identification of Kodoko virus, a novel arenavirus of the African pigmy mouse (Mus Nannomys minutoides) in West Africa. Virology. 2007;364:178–83. DOIPubMedGoogle Scholar
- van der Groen G, Kurata T, Mets C. Modifications to indirect immunofluorescence tests on Lassa, Marburg, and Ebola material. Lancet. 1983;1:654. DOIPubMedGoogle Scholar
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64. DOIPubMedGoogle Scholar
- Günther S, Emmerich P, Laue T, Kühle O, Asper M, Jung A, Imported Lassa fever in Germany: molecular characterization of a new Lassa virus strain. Emerg Infect Dis. 2000;6:466–76. DOIPubMedGoogle Scholar
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. DOIPubMedGoogle Scholar
- Essbauer SS, Schmidt-Chanasit J, Madeja EL, Wegener W, Friedrich R, Petraityte R, Nephropathia epidemica in metropolitan area, Germany. Emerg Infect Dis. 2007;13:1271–3.PubMedGoogle Scholar
- Walker DH, Wulff H, Lange JV, Murphy FA. Comparative pathology of Lassa virus infection in monkeys, guinea-pigs, and Mastomys natalensis. Bull World Health Organ. 1975;52:523–34.PubMedGoogle Scholar
- Vieth S, Torda AE, Asper M, Schmitz H, Günther S. Sequence analysis of L RNA of Lassa virus. Virology. 2004;318:153–68. DOIPubMedGoogle Scholar
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–65. DOIPubMedGoogle Scholar