Volume 15, Number 2—February 2009
Research
Identification of Melioidosis Outbreak by Multilocus Variable Number Tandem Repeat Analysis
Figure 1
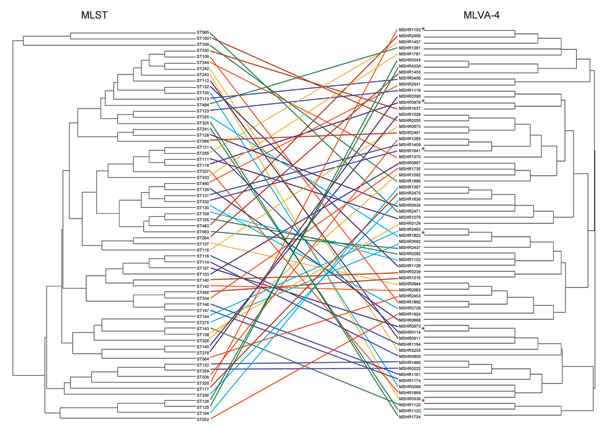
Figure 1. Comparison of multilocus sequence typing (MLST) and 4-locus multilocus variable number tandem repeat analysis (MLVA-4) dendrograms for 65 Burkholderia pseudomallei isolates. MLST sequence type (ST) is shown for each isolate, with the corresponding isolate number listed for the MLVA-4 profile and shown by the colored lines. The red asterisks indicate 6 isolates that represent diversity of MLVA-4; these isolates were used to calibrate the dendrogram in Figure 3.
Page created: December 08, 2010
Page updated: December 08, 2010
Page reviewed: December 08, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.