Volume 15, Number 5—May 2009
Research
Cross-Border Dissemination of Methicillin-Resistant Staphylococcus aureus, Euregio Meuse-Rhin Region
Abstract
Because the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) differs among the 3 countries forming the Euregio Meuse-Rhin (EMR) region (Belgium, Germany, and the Netherlands), cross-border healthcare requires information about the spread of MRSA in the EMR. We investigated the emergence, dissemination, and diversity of MRSA clones in the EMR by using several typing methods. MRSA associated with clonal complexes 5, 8, 30, and 45 was disseminated throughout the EMR. Dutch isolates, mainly associated with sequence types (ST) ST5-MRSA-II, ST5-MRSA-IV, ST8-MRSA-IV, and ST45-MSRA-IV had a more diverse genetic background than the isolates from Belgium and Germany, associated with ST45-MRSA-IV and ST5-MRSA-II, respectively. MRSA associated with pigs (ST398-MRSA-IV/V) was found in the Dutch area of the EMR. Five percent of the MRSA isolates harbored Panton-Valentine leukocidin and were classified as community-associated MRSA associated with ST1, 8, 30, 80, and 89.
Almost one third of the European population lives in a border region (Euregio). These border regions have collaborated since the late 1950s, especially in the field of healthcare (1). Cross-border patient mobility and free access to healthcare facilities within the European Union in general, and the Euregios in particular, are important for patients, medical doctors, healthcare facilities, and healthcare insurance companies. The Euregio Meuse-Rhin (EMR), an area totaling 4,973 square miles (12,882 km2), is the border region of Belgium, Germany, and the Netherlands. The EMR comprises the Belgian provinces of Limburg and Liège, the German-speaking region of Belgium, the Aachen region in Germany, and the southern part of the Dutch province of Limburg. Each year, thousands of the 3.88 million inhabitants of the EMR cross the border to consult a medical specialist or a healthcare facility. Since 2003, hospitals in the EMR have built a strong collaboration. For example, the University Hospital Maastricht in the Netherlands and the University Hospital Aachen in Germany have an official agreement for the transfer of patients; consequently, dozens of patients are transferred each year between the 2 hospitals. The same applies for the University Hospital Maastricht in the Netherlands and the General Hospital Vesalius in Belgium, between which nearly 100 patients are transferred each year. In an official publication of the European Commission (D. Byrne, Maastricht Conference on Cross-Border Health Care, Maastricht, the Netherlands, June 8, 2004), the EMR was mentioned as a model region for the European Union in the field of cross-border healthcare and cross-border cooperation of hospitals. Furthermore, in July 2008, establishment of a pan-European university hospital was announced, a collaboration among the university hospitals of Maastricht in the Netherlands and Aachen in Germany.
Of particular concern is cross-border dissemination of multidrug-resistant bacteria, such as methicillin-resistant Staphylococcus aureus (MRSA). The 3 countries forming the EMR differ considerably in the prevalence of hospital-isolated MRSA (23.6%, 13.8%, and 0.6% in Belgium, Germany, and the Netherlands, respectively) (2). Consequently, cross-border transfer of patients may affect the dissemination and prevalence of MRSA, particularly when patients are transferred from countries with a relatively high prevalence to a country with a low prevalence.
A study of MRSA isolates from the EMR between December 1999 and February 2004 showed that isolates from clonal complex (CC) 5 and CC 8, which harbor the resistance elements staphylococcal cassette chromosome mec (SCCmec) types I–IV, had been disseminated in the EMR (2). Our aim was to investigate the emergence, dissemination, and diversity of MRSA clones in the EMR during a 10-month period in 2005 and 2006 and to compare the results with those of the previous study. We used sequencing of the short sequence repeat (SSR) region of the S. aureus protein A gene (spa typing), multilocus sequence typing (MLST), and SCCmec typing by PCR to investigate the genetic background of all MRSA isolates. The spa locus was typed to provide more detailed information about prevalent MRSA clones in the EMR, especially because the previous study used only MLST analyses on a small subset of isolates (2). Finally, because an increase of Panton-Valentine leukocidin (PVL)–positive MRSA isolates in the Netherlands has recently been observed (3), we investigated the possible spread of PVL-positive MRSA clones into hospitals in the EMR, as well as the prevalence of the virulence factors collagen adhesion (CNA) and toxic shock syndrome toxin-1 (TSST-1).
MRSA Isolates
We investigated 257 MRSA isolates, cultured during July 2005–April 2006 from 8 geographically closely related hospitals in the EMR. The hospitals included 1 hospital in Belgium (General Hospital Vesalius, Tongeren, 355 beds), 2 hospitals in Germany (General Hospital Dūren, 521 beds, and Marien Hospital, Aachen, 321 beds), and 5 hospitals in the Netherlands (Atrium Medical Center, Heerlen, 811 beds; Orbis Medical and Care Center, Sittard, 578 beds; Laurentius Hospital, Roermond, a 458-bed general hospital; University Hospital Maastricht, a tertiary hospital, 680 beds; and VieCuri Medical Center, Venlo, a 554-bed general hospital). The 257 MRSA isolates comprised 44 from Belgium, 92 from Germany, and 121 from the Netherlands. Isolates from the Belgian and German hospitals were from patients with MRSA infection; Dutch isolates were from patients carrying MRSA who were admitted to the Dutch hospitals. All isolates were identified as S. aureus by Gram stain, catalase, and coagulase testing. The presence of the mecA gene was determined as described previously (2).
Antimicrobial Drug Susceptibility Testing
The susceptibility pattern of the MRSA isolates was determined according to the guidelines of the Clinical and Laboratory Standards Institute (4). Susceptibility to the following antimicrobial agents was determined as MIC: cefaclor, cefuroxime, clindamycin, ciprofloxacin, clarithromycin, gentamicin, linezolid, moxifloxacin, oxacillin, penicillin, rifampin, teicoplanin, tetracycline, trimethoprim/sulfamethoxazole, and vancomycin. The susceptibility to fucidic acid and mupirocin (Rosco, Taastrup, Denmark) was determined by using the disk-diffusion method (5,6). MRSA isolates resistant to clarithromycin were tested for inducible clindamycin resistance by using the D-test (7).
Typing Methods
SCCmec typing was performed as described by Oliveira et al. (8) with the modification described previously (2). SCCmec type I elements that lack locus A (pls region) are indistinguishable (9) from SCCmec type IV elements when the method of Oliveira et al. is used (8). Furthermore, locus D (dcs region) is detected in both SCCmec types IV and VI (10). Therefore, SCCmec elements that were typed as SCCmec type IV using the method of Oliveira et al. (8) were further analyzed for presence of the ccrAB gene. SCCmec elements that could not be typed with the method of Oliveira et al (8) were further analyzed by using the methods of Ito et al. (11) and Zhang et al. (12).
Real-time amplification of the spa gene was performed as described previously, followed by sequencing of the SSR region (13). The spa types were clustered into spa-CCs using the algorithm Based Upon Repeat Pattern (BURP) with the Ridom StaphType version 1.4 software package (www.ridom.de). Because spa typing, together with the algorithm BURP, yields results concordant with typing results obtained by MLST and pulsed-field gel electrophoresis (13), the associated CCs, as determined with MLST, were allocated through the Ridom SpaServer (http://spaserver.ridom.de). To confirm the association between MLST and spa typing, in combination with BURP, MLST was performed on a representative set of 12 strains of each major spa type and spa-CC (2).The presence of CNA, PVL, and TSST-1 was determined with real-time PCR assays (14,15).
Antimicrobial Drug Susceptibility Patterns
All 257 MRSA isolates were resistant to the β-lactam antimicrobial agents cefaclor, cefuroxime, oxacillin, and penicillin and were susceptible to linezolid, teicoplanin, and vancomycin. Most isolates were also resistant to ciprofloxacin (84%) and moxifloxacin (82%). The Dutch MRSA isolates were more often susceptible to ciprofloxacin and moxifloxacin than were the Belgian and German isolates (Table 1) (p<0.05). Furthermore, 78% of the MRSA isolates were resistant to clarithromycin, and 62%, to clindamycin. Susceptibility for clarithromycin and clindamycin differed by country (Table 1). A total of 41 MRSA isolates (19 from Belgium, 5 from Germany, and 17 from the Netherlands) was resistant to clarithromycin and susceptible to clindamycin. The D-test showed that 31 (76%) of these 41 MRSA isolates had the inducible clindamycin resistant phenotype, including 15 from Belgium, 5 from Germany, and 11 from the Netherlands.
Distribution of MRSA Clones
SCCmec type IV was predominant in MRSA isolates from Belgium (77%), whereas MRSA isolates from Germany harbored mainly SCCmec type II (82%). MRSA isolates from the Dutch region harbored both SCCmec type II and IV (27% and 65%, respectively). Although 25 (10%) of the 257 MRSA isolates harbored an SCCmec element that could not be typed with the method of Oliveira et al. (8), they could be typed with the other methods. Seven MRSA isolates from Belgium harbored a SCCmec type III element that lacked Tn554, which is usually characteristic for SCCmec type III. From the German region, 1 isolate that had a nontypeable SCCmec element harbored ccrC, locus E, and Tn554. The method of Zhang et al. (12) classified this element as SCCmec type III. In the Netherlands, 17 MRSA isolates contained a nontypeable SCCmec element as defined by Oliveira et al. (8). Ten of these were classified as SCCmec type IV, lacking locus D. The remaining 7 harbored ccrC, characteristic for SCCmec type V, and were classified as such with the method of Zhang et al. (12).
The 257 MRSA isolates were classified into 36 different spa types, and BURP analysis showed 6 spa-CCs, 4 singletons, and 3 spa types that were excluded from the analysis because the spa region was <5 spa repeats long (Table 2). MLST analyses showed 10 different STs among the 12 MRSA strains (Table 2). In the EMR, spa-CC 045 (MLST CC5; 21%) and spa-CC 038 (MLST CC45; 75%) were found predominantly among MRSA isolates from the Belgian region; spa-CC 045 (MLST CC5; 85%) was found among isolates from the German region. The Dutch MRSA isolates were grouped into spa-CC 045 (MLST CC5; 39%), spa-CC 019/012/318/011/108 (MLST CC30 and CC398; 15%), spa-CC 038 (MLST CC45; 15%), spa-CC with no founder 5 (MLST CC8; 16%), and spa-CC with no founder 6 (MLST CC 45; 5%).
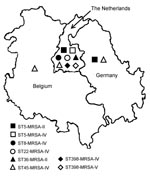
The ST5-MRSA-II (New York/Japan) clone was found mainly in Germany and the Netherlands, and the ST45-MRSA-IV (Berlin) clone was found in Belgium and the Netherlands. Furthermore, the ST5-MRSA-IV (Pediatric) clone was found among the Dutch isolates. The MRSA isolates classified as CC30 (ST30-MRSA-IV and ST36-MRSA-II) were found only in the Netherlands. Most of the ST8-MRSA-IV (UK EMRSA-2/6) isolates were found in the Netherlands. Furthermore, several ST398-MRSA-IV and ST398-MRSA-V isolates were found in certain Dutch hospitals (Figure, Table 3).
Prevalence of Virulence Factors
Eleven (5%) of the 257 MRSA isolates were PVL-positive. These isolates were associated with different genetic backgrounds, i.e., ST1-MRSA-V (1 Dutch isolate), ST8-MRSA-IV, ST30-MRSA-IV (2 Dutch isolates each), ST45-MRSA-IV (1 isolate from Germany), ST80-MRSA-IV (1 isolate from Germany and 2 from the Netherlands), ST89-MRSA-IV and ST89-MRSA-V (1 Dutch isolate each). Six of the PVL-positive isolates were positive for the cna gene, and none harbored the tst gene.
Nine (4%) of the 257 MRSA isolates were positive for the tst gene, 4 isolates were classified as ST22-MRSA-IV, 3 as ST36-MRSA-II, 1 as ST30-MRSA-IV, and 1 could not be classified as an MRSA clone (spa type t779). All isolates were from the Netherlands and were positive for the cna gene; none harbored PVL.
Ninety-five (37%) of the 257 MRSA isolates were positive for the cna gene (34 from Belgium, 9 from Germany, and 52 from the Netherlands). All MRSA isolates from CC30, CC45, and ST398 harbored the cna gene. Furthermore, 1 isolate from CC5, 1 from CC80, 6 classified as singletons (associated with ST22 and ST89), and 2 excluded from the BURP analyses were positive for the cna gene.
Because cross-border healthcare is an issue in the EMR, and the prevalence of MRSA differs among the countries forming the EMR, studying the possible emergence, spread, and diversity of MRSA clones within and among these countries is important (2). In addition to MRSA clones from CC5 and CC8, found previously in the EMR, we observed MRSA isolates from CC30 and CC45. Furthermore, the Dutch isolates had a more heterogeneous genetic background than did MRSA isolates from Belgium and Germany. The prevalence of PVL-positive MRSA isolates, belonging to ST1, 8, 30, 80 and 89, was higher than that found in the previous study (5% vs. 1.3%) (2).
The antimicrobial susceptibility of the MRSA isolates depends on the S. aureus lineage. The observation that the Dutch MRSA isolates were more often susceptible to ciprofloxacin and moxifloxacin than were isolates from Belgium and Germany can be explained by the fact that the isolates associated with ST5-MRSA-IV, ST22-MRSA-IV, and ST30-MRSA-IV, which were susceptible to ciprofloxacin and moxifloxacin, were mainly observed in the Netherlands. Although ST22-MRSA-IV is commonly susceptible to tetracycline, the ST22-MRSA-IV isolates in this study were resistant to tetracycline (16). S. aureus can harbor resistance genes on mobile genetic elements on the genome, such as Tn554, as well as on plasmids, and these can be exchanged among S. aureus lineages, possibly because of antimicrobial drug pressure (17).
Primarily because of the Dutch “search-and-destroy” policy, isolates derived from colonized persons were available from the Netherlands, whereas isolates from Belgium and Germany were derived from infections. However, nasal carriers are at increased risk of acquiring MRSA infection (18). Consequently, not preventing the spread of MRSA among nasal carriers could lead to MRSA infection among these persons. Furthermore, the molecular epidemiology of MRSA can vary widely among hospitals. In the Dutch hospitals of the EMR, MRSA clones in each hospital were diverse, whereas in the Belgian hospital and 2 German hospitals, 1 MRSA clone predominated, showing that the number of hospitals is unlikely to have biased the results of our study.
Most of the MRSA isolates from Belgium were associated with the Berlin clone (ST45-MRSA-IV). This clone has previously been found in Belgium, Germany, and the Netherlands (19). Most of the MRSA isolates from Germany were associated with the New York/Japan clone (ST5-MRSA-II), previously found in Belgium and Germany (2,19). Most of the Dutch MRSA isolates belonged to 5 MRSA clones (Table 3). Twenty-five percent of the Dutch isolates were associated with the New York/Japan clone (ST5-MRSA-II), which has not been previously found in the Netherlands. The Pediatric clone (ST5-MRSA-IV), which represented 14% of the Dutch isolates, has been found in Belgium but not in the Netherlands (20,21). The Berlin clone (ST45-MRSA-IV), comprising 21% of the Dutch isolates, and the UK EMRSA-2/-6 clone (ST8-MRSA-IV), comprising 16% of the Dutch isolates, have been described in all 3 EMR countries (19,20). In addition, some less prevalent MRSA clones were observed. Four tst-positive MRSA isolates were associated with the UK EMRSA-15 clone (ST22-MRSA-IV), previously found in Belgium and Germany but not in the Netherlands (19,20). Three Dutch MRSA isolates (spa type t012), harboring SCCmec type II, were associated with the CC30 lineage. These isolates might be derived from the UK EMRSA-16 (ST36-MRSA-II) clone (spa type t018) because spa types t012 and t018 differ in 1 spa repeat (r24) and are thus related. Furthermore, both clones harbor the cna and tst genes (22,23). The highly endemic UK EMRSA-16 clone has not been observed before in the Netherlands, although this clone has previously been found in Belgium (24). Seven and 5 of the Dutch MRSA isolates were associated with ST398-MRSA-IV and ST398-MRSA-V, respectively, MRSA clones usually observed in pigs and among screening samples from pig farmers (25). The ST398 clone is now observed among screening samples of veterinarians from many countries in Europe, including Belgium and Germany (26). However, ST398 also has been isolated from several forms of human infections in Germany (27). The ST398 isolates from our study were positive for the cna gene, suggesting a higher virulence than that of the CNA-negative German ST398 MRSA isolates (27). One Dutch MRSA strain was associated with the ST30-MRSA-IV clone, previously reported in Belgium, Germany, and France (20,21,28). The more diverse genetic background among MRSA isolates in the Dutch part of the EMR and the close cooperation of hospitals in the EMR might suggest that importation of MRSA from Belgium and Germany has occurred through cross-border healthcare (Table 4) (2). Other, less likely, explanations for the diversity of MRSA clones in the Netherlands are the spread of MRSA from countries other than Belgium or Germany (19) and the emergence of new MRSA clones in vivo through transfer of the SCCmec element from methicillin-resistant coagulase-negative staphylococci to methicillin-sensitive S. aureus strains (29).
We could not determine the SCCmec type for 10% of the MRSA isolates by using the method of Oliveira et al. (8). This percentage was similar to that found in other studies (30,31) but higher than the 3% previously found in the EMR (2). The relatively large number of nontypeable SCCmec types found in this study, probably formed by homologous recombination among SCCmec elements, supports the need for a new system for SCCmec typing and nomenclature (19).
The 7 Belgian MRSA isolates with the nontypeable SCCmec type III element were associated with CC5 and had the related spa types t045 and t1107 (http://spaserver.ridom.de). Although SCCmec type III usually is found in the CC8 genetic background, such as in the ST239-MRSA-III clone, an MRSA associated with CC5 (spa type t045) and harboring SCCmec type III recently was observed in Belgium (32). This might suggest that a new MRSA clone, ST5-MRSA-III, is beginning to emerge in Belgium.
The nontypeable SCCmec element of the German MRSA isolate harbored locus E and ccrC, specific for SCCmec type V (2), and Tn554, normally carried by SCCmec type II, III, and SCCmercury. Zhang et al. (12) classified this element as SCCmec type III, but the SCCmec type III-specific primers used by this method are situated near locus E on SCCmercury (33), indicating that this element could be a SCCmercury element containing mecA. Further investigation is needed into the structure of this element.
Previous studies have shown that MRSA isolates classified as community-associated usually harbor either SCCmec type IV or V, and often PVL, but may differ in their genetic backgrounds (CC1, CC8, CC30, CC59 and CC80) (34). In the EMR, 5% of the MRSA isolates were positive for PVL, which is higher than the previously reported 1.3% (2). Thus, PVL-positive MRSA isolates with a heterogeneous genetic background are emerging in the EMR.
PVL-positive MRSA isolates associated with ST8-MRSA-IV, ST30-MRSA-IV, and ST80-MRSA-IV have been isolated in the Netherlands (3,35). In the present study, 2 of the PVL-positive MRSA isolates harbored SCCmec type V. The different genetic background of these isolates, i.e., ST89 and ST772, a single-locus variant of ST1 at the pta locus, might suggest that SCCmec type V was introduced on different occasions into different S. aureus lineages. A PVL-positive ST772-MRSA-V has been observed in Germany (36). One of the PVL-positive isolates harbored SCCmec type I, and such isolates with a ST30 and ST37 genetic background have been described in the Netherlands (3). Although a recent study suggested that CNA and PVL combined contribute to virulence, only 6 of the 11 PVL-positive MRSA isolates from the EMR harbored the cna gene (37). Further studies are needed to investigate the contribution of the combination of CNA and PVL to virulence.
The genetic background of 1 PVL-positive ST45-MRSA-IV isolate from Belgium was similar to that of the Berlin clone. Hitherto, only PVL-negative isolates with this background have been found in EMR countries (19,20). PVL-positive MRSA isolates, associated with the major CA-MRSA clones (ST8-MRSA-IV, ST30-MRSA-IV, and ST80-MRSA-IV) have been reported from Belgium (38). Because PVL is situated on a phage, the genes encoding PVL might have been transferred to S. aureus with a CC45 genetic background (34).
Our study found a PVL-positive MRSA isolate from Germany with spa type t042 (spa repeat pattern r26r23r12r34r34r33r34). This spa type is strongly related to spa types t044 and t131 (spa repeat patterns r07r23r12r34r34r33r34 and r07r23r12r34r33r34, respectively), which are usually associated with the CA-MRSA ST80-MRSA-IV clone found in Germany (34).
The cna gene has been previously observed among MRSA isolates from CC22, CC30, and CC45 (23,29). Therefore, the presence of the can gene might, together with spa typing, be used as a marker for different genetic backgrounds.
MRSA clones associated with the hospital associated-MRSA CCs 5, 8, 22, 30, and 45, the PVL-positive CA-MRSA CCs 1, 8, 30, 80, and 89, as well as MRSA related to pigs (ST389-MRSA-IV/V) were observed in the EMR. Dissemination of these clones is possible because of the introduction of new MRSA clones associated with travel; with patients who have previously been admitted to a hospital abroad (cross-border healthcare); or with other high-risk patients, such as pig-farmers and their families. Therefore, a cross-border search-and-contain policy may help control the further spread of MRSA and reduce the financial cost to hospitals, nursing homes, and the community in the EMR.
Dr Deurenberg is a postdoctoral fellow at the Department of Medical Microbiology at the University Hospital Maastricht. His research interests focus on molecular diagnostics, mechanisms of antimicrobial drug resistance, and the molecular epidemiology of S. aureus.
Acknowledgment
The study was partly performed within the framework of the Interreg-III project “Cross-Border Health Care in the Euregio Meuse-Rhin.”
References
- Wolf U, Hollederer A, Brand H. Cross-border cooperation in Europe: what are Euregios? Gesundheitswesen. 2006;68:667–73. DOIPubMedGoogle Scholar
- Deurenberg RH, Vink C, Oudhuis GJ, Mooij JE, Driessen C, Coppens G, Different clonal complexes of methicillin-resistant Staphylococcus aureus are disseminated in the Euregio Meuse-Rhine region. Antimicrob Agents Chemother. 2005;49:4263–71. DOIPubMedGoogle Scholar
- Wannet WJ, Spalburg E, Heck ME, Pluister GN, Tiemersma E, Willems RJ, Emergence of virulent methicillin-resistant Staphylococcus aureus strains carrying Panton-Valentine leukocidin genes in the Netherlands. J Clin Microbiol. 2005;43:3341–5. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, Twelfth informational supplement, M100-S12. Wayne (PA): The Institute; 2002.
- Toma E, Barriault D. Antimicrobial activity of fusidic acid and disk diffusion susceptibility testing criteria for gram-positive cocci. J Clin Microbiol. 1995;33:1712–5.PubMedGoogle Scholar
- Fuchs PC, Jones RN, Barry AL. Interpretive criteria for disk diffusion susceptibility testing of mupirocin, a topical antibiotic. J Clin Microbiol. 1990;28:608–9.PubMedGoogle Scholar
- O’Sullivan MV, Cai Y, Kong F, Zeng X, Gilbert GL. Influence of disk separation distance on accuracy of the disk approximation test for detection of inducible clindamycin resistance in Staphylococcus spp. J Clin Microbiol. 2006;44:4072–6. DOIPubMedGoogle Scholar
- Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–61. DOIPubMedGoogle Scholar
- Shore A, Rossney AS, Keane CT, Enright MC, Coleman DC. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob Agents Chemother. 2005;49:2070–83. DOIPubMedGoogle Scholar
- Oliveira DC, Milheirico C, de Lencastre H. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob Agents Chemother. 2006;50:3457–9. DOIPubMedGoogle Scholar
- Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 2004;48:2637–51. DOIPubMedGoogle Scholar
- Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–33. DOIPubMedGoogle Scholar
- Nulens E, Stobberingh EE, van Dessel H, Sebastian S, van Tiel FH, Beisser PS, Molecular characterization of Staphylococcus aureus bloodstream isolates collected in a Dutch university hospital between 1999 and 2006. J Clin Microbiol. 2008;46:2438–41. DOIPubMedGoogle Scholar
- Deurenberg RH, Vink C, Driessen C, Bes M, London N, Etienne J, Rapid detection of Panton-Valentine leukocidin from clinical isolates of Staphylococcus aureus strains by real-time PCR. FEMS Microbiol Lett. 2004;240:225–8. DOIPubMedGoogle Scholar
- Deurenberg RH, Nieuwenhuis RF, Driessen C, London N, Stassen FR, van Tiel FH, The prevalence of the Staphylococcus aureus tst gene among community- and hospital-acquired strains and isolates from Wegener's granulomatosis patients. FEMS Microbiol Lett. 2005;245:185–9. DOIPubMedGoogle Scholar
- Amorim ML, Faria NA, Oliveira DC, Vasconcelos C, Cabeda JC, Mendes AC, Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J Clin Microbiol. 2007;45:2881–8. DOIPubMedGoogle Scholar
- Lindsay JA, Holden MT. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct Integr Genomics. 2006;6:186–201. DOIPubMedGoogle Scholar
- Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–62. DOIPubMedGoogle Scholar
- Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8:747–63. DOIPubMedGoogle Scholar
- Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A. 2002;99:7687–92. DOIPubMedGoogle Scholar
- Hallin M, Denis O, Deplano A, De Mendonca R, De Ryck R, Rottiers S, Genetic relatedness between methicillin-susceptible and methicillin-resistant Staphylococcus aureus: results of a national survey. J Antimicrob Chemother. 2007;59:465–72. DOIPubMedGoogle Scholar
- Ferry T, Bes M, Dauwalder O, Meugnier H, Lina G, Forey F, Toxin gene content of the Lyon methicillin-resistant Staphylococcus aureus clone compared with that of other pandemic clones. J Clin Microbiol. 2006;44:2642–4. DOIPubMedGoogle Scholar
- Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2006;193:1495–503. DOIPubMedGoogle Scholar
- Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003;41:1574–85. DOIPubMedGoogle Scholar
- van Rijen MM, Van Keulen PH, Kluytmans JA. Increase in a Dutch hospital of methicillin-resistant Staphylococcus aureus related to animal farming. Clin Infect Dis. 2008;46:261–3. DOIPubMedGoogle Scholar
- Wulf MW, Sorum M, van Nes A, Skov R, Melchers WJ, Klaassen CH, Prevalence of methicillin-resistant Staphylococcus aureus among veterinarians: an international study. Clin Microbiol Infect. 2008;14:29–34. DOIPubMedGoogle Scholar
- Witte W, Strommenger B, Stanek C, Cuny C. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg Infect Dis. 2007;13:255–8.PubMedGoogle Scholar
- Durand G, Bes M, Meugnier H, Enright MC, Forey F, Liassine N, Detection of new methicillin-resistant Staphylococcus aureus clones containing the toxic shock syndrome toxin 1 gene responsible for hospital- and community-acquired infections in France. J Clin Microbiol. 2006;44:847–53. DOIPubMedGoogle Scholar
- Wielders CL, Vriens MR, Brisse S, de Graaf-Miltenburg LA, Troelstra A, Fleer A, In-vivo transfer of mecA DNA to Staphylococcus aureus. Lancet. 2001;357:1674–5. DOIPubMedGoogle Scholar
- Hanssen AM, Kjeldsen G, Sollid JU. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob Agents Chemother. 2004;48:285–96. DOIPubMedGoogle Scholar
- Chung M, Dickinson G, De Lencastre H, Tomasz A. International clones of methicillin-resistant Staphylococcus aureus in two hospitals in Miami, Florida. J Clin Microbiol. 2004;42:542–7. DOIPubMedGoogle Scholar
- Deplano A, De Mendonca R, De Ryck R, Struelens MJ. External quality assessment of molecular typing of Staphylococcus aureus isolates by a network of laboratories. J Clin Microbiol. 2006;44:3236–44. DOIPubMedGoogle Scholar
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–74. DOIPubMedGoogle Scholar
- Tristan A, Bes M, Meugnier H, Lina G, Bozdogan B, Courvalin P, Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis. 2007;13:594–600.PubMedGoogle Scholar
- Huijsdens XW, van Santen-Verheuvel MG, Spalburg E, Heck ME, Pluister GN, Eijkelkamp BA, Multiple cases of familial transmission of community-acquired methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2006;44:2994–6. DOIPubMedGoogle Scholar
- Strommenger B, Braulke C, Pasemann B, Schmidt C, Witte W. Multiplex PCR for rapid detection of Staphylococcus aureus isolates suspected to represent community-acquired strains. J Clin Microbiol. 2008;46:582–7. DOIPubMedGoogle Scholar
- de Bentzmann S, Tristan A, Etienne J, Brousse N, Vandenesch F, Lina G. Staphylococcus aureus isolates associated with necrotizing pneumonia bind to basement membrane type I and IV collagens and laminin. J Infect Dis. 2004;190:1506–15. DOIPubMedGoogle Scholar
- Denis O, Deplano A, De Beenhouwer H, Hallin M, Huysmans G, Garrino MG, Polyclonal emergence and importation of community-acquired methicillin-resistant Staphylococcus aureus strains harbouring Panton-Valentine leukocidin genes in Belgium. J Antimicrob Chemother. 2005;56:1103–6. DOIPubMedGoogle Scholar
- Lindsay JA, Moore CE, Day NP, Peacock SJ, Witney AA, Stabler RA, Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J Bacteriol. 2006;188:669–76. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 15, Number 5—May 2009
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Ruud H. Deurenberg, Department of Medical Microbiology, Maastricht Infection Center, University Hospital Maastricht, PO Box 5800, 6202 AZ Maastricht, the Netherlands
Top