Volume 16, Number 4—April 2010
Letter
Retraction: Triple Reassortant Swine Influenza A (H3N2) Virus in Waterfowl
Figure
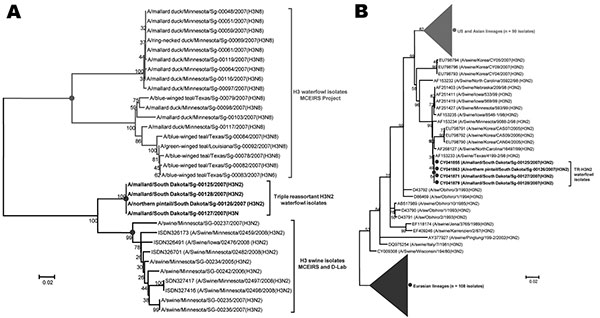
Figure. Phylogenetic analysis of hemagglutinin (HA) sequences from waterfowl strains isolated in this study (boldface), based on the HA gene sequences. The evolutionary associations were inferred in MEGA4.0 (www.megasoftware.net) by using the neighbor-joining algorithm with the Kimura 2-parameter gamma model and 1,000 bootstrap replications (shown on branch bifurcations). A) Evolutionary distances of waterfowl isolates from swine and avian HA (H3) sequences from the Minnesota Center of Excellence for Influenza Research and Surveillance (MCEIRS) sequencing project or Minnesota Veterinary Diagnostic Laboratory (D-Lab) database. B) Phylogeny of 230 strains, including Eurasian and North American lineages of influenza A (H3N2) viruses. Data suggest swine influenza virus (H3N2) ancestry in the waterfowl strains. GenBank accession numbers are shown. Scale bars indicate nucleotide substitutions per site.