Volume 16, Number 6—June 2010
Dispatch
Transfer of Carbapenem-Resistant Plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in Patient
Figure 1
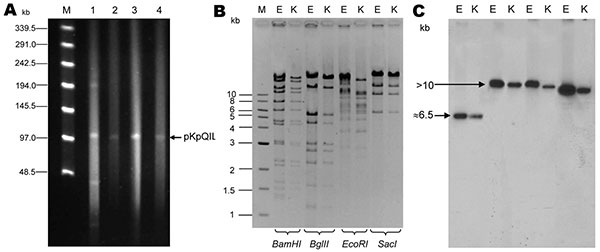
Figure 1. A) Analysis of Klebsiella pneumoniae carbapenemase (KPC)–encoding plasmids in isolates Kpn1 (1), Eco2 (3), Kpn1-T (2), and Eco2-T (4), Israel, 2008. Plasmid size estimation was performed by digestion of DNA with S1 nuclease (20 U; Promega, Madison, WI, USA) followed by pulsed-field gel electrophoresis (PFGE) with the CHEF-DR III apparatus (Bio-Rad Laboratories, Inc., Hercules, CA, USA), as described (8–11). Lambda ladder PFG marker (New England Biolabs, Beverly, MA, USA) was used as a molecular size marker (lane M). B) Restriction fragment length polymorphism of the KPC-3–encoding plasmid from Kpn1-T (K) and Eco2-T (E). Plasmid DNA was digested with BamHI, BglII, EcoRI, and SacI endonucleases (New England Biolabs) and underwent PFGE on a 1% agarose gel. The level of similarity between restriction patterns was calculated by using GelcomparII software version 5 (Applied Maths, Kortrigk, Belgium). Lane 1, 1-kb DNA ladder (New England Biolabs). C) Southern blot analysis of plasmid DNA hybridized with blaKPC-3-labeled probe. Plasmid restriction products were transferred to a Hybond N+ membrane (Amersham Biosciences, Little Chalfont, United Kingdom), cross-linked with UV light, and hybridized with a blaKPC-3-labeled probe (892-bp product of blaKPC-3).
References
- Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. DOIPubMedGoogle Scholar
- Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53:3365–70. DOIPubMedGoogle Scholar
- Woodford N, Zhang J, Warner M, Kaufmann ME, Matos J, Macdonald A, et al. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J Antimicrob Chemother. 2008;62:1261–4. DOIPubMedGoogle Scholar
- Leavitt A, Navon-Venezia S, Chmelnitsky I, Schwaber MJ, Carmeli Y. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob Agents Chemother. 2007;51:3026–9. DOIPubMedGoogle Scholar
- Schwaber MJ, Klarfeld-Lidji S, Navon-Venezia S, Schwartz D, Leavitt A, Carmeli Y. Antimicrob Agents Chemother. 2008;52:1028–33.Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. DOIPubMedGoogle Scholar
- Schwaber MJ, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: a potential threat.JAMA. 2008;300:2911–3.
- Navon-Venezia S, Chmelnitsky I, Leavitt A, Schwaber MJ, Schwartz D, Carmeli Y. Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob Agents Chemother. 2006;50:3098–101. DOIPubMedGoogle Scholar
- Noller AC, McEllistrem MC, Stine OC, Morris JG Jr, Boxrud DJ, Dixon B, et al. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J Clin Microbiol. 2003;41:675–9. DOIPubMedGoogle Scholar
- Schlesinger J, Navon-Venezia S, Chmelnitsky I, Hammer-Münz O, Leavitt A, Gold HS, et al. Extended-spectrum beta-lactamases among Enterobacter isolates obtained in Tel Aviv, Israel. Antimicrob Agents Chemother. 2005;49:1150–6. DOIPubMedGoogle Scholar
- Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226:235–40. DOIPubMedGoogle Scholar
- Li L, Lim CK. A novel large plasmid carrying multiple beta-lactam resistance genes isolated from a Klebsiella pneumoniae strain. J Appl Microbiol. 2000;88:1038–48. DOIPubMedGoogle Scholar
- Navon-Venezia S, Leavitt A, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, et al. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob Agents Chemother. 2009;53:818–20. DOIPubMedGoogle Scholar
- Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. Plasmid pKpQIL harboring KPC-3 and TEM-1 renders carbapenem resistance in extremely drug resistant epidemic Klebsiella pneumoniae. J Antimicrob Chemother. 2009. In press.PubMedGoogle Scholar
- Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob Agents Chemother. 2008;52:1257–63. DOIPubMedGoogle Scholar
- Wielders CL, Vriens MR, Brisse S, de Graaf-Miltenburg LA, Troelstra A, Fleer A, Evidence for in-vivo transfer of mecA DNA between strains of Staphylococcus aureus. Lancet. 2001;357:1674–5. DOIPubMedGoogle Scholar