Volume 16, Number 6—June 2010
Dispatch
Rhinovirus C and Respiratory Exacerbations in Children with Cystic Fibrosis
Cite This Article
Citation for Media
Abstract
To investigate a possible role for human rhinovirus C in respiratory exacerbations of children with cystic fibrosis, we conducted microbiologic testing on respiratory specimens from 103 such patients in São Paulo, Brazil, during 2006–2007. A significant association was found between the presence of human rhinovirus C and respiratory exacerbations.
Cystic fibrosis (CF) is an autosomal inherited disease characterized by recurrent and chronic respiratory infections that ultimately lead to the need for a lung transplant early in life or to death (1). The role of bacterial infections in CF is well established, and most treatments focus on eradication or suppression of bacterial infections (mainly those caused by Pseudomonas aeruginosa) (1).
Respiratory viruses such as respiratory syncytial virus (RSV) and influenza also seem to cause early damage or increase the risk for respiratory exacerbations (2,3) in these patients. However, the role of newly described respiratory viruses is not well known. Infection of these patients with human rhinovirus (HRV), a member of the family Picornaviridae, has been described. Although some studies have suggested a substantial pathogenic role for these viruses (3), controversy still exists (4).
Recently a new clade of human rhinovirus, named rhinovirus C (5), was identified through molecular methods. This new clade has been found throughout the world (6), and some studies have attributed severe respiratory infections in children to this agent (7,8). We investigated whether this agent played a role in the respiratory exacerbations of children and adolescents with CF who attended the Instituto da Crinaça, Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo.
A total of 103 CF patients (49 girls, 54 boys; median age 8.9 years; age range 3.8 months–17.8 years) were enrolled in the study from September 6, 2006 through September 4, 2007. Nasopharyngeal aspirates or nasal mucus specimens for viral investigation, as well as sputum or oropharyngeal samples for microbiology culture, were collected during scheduled visits or unscheduled visits on 408 occasions, with a median ± SD of 4 ± 1.74 visits per patient (range 1–9 visits).
Clinical and lung function data were obtained at all visits. Exacerbation of respiratory disease was defined as the presence of >2 of the following signs or symptoms: fever, increase in the amount of secretion or cough intensity, change in sputum’s color, worsening of dyspnea, loss of appetite, a decrease of forced expiratory volume in 1 s >10%, and weight loss.
Total nucleic acids were extracted from nasopharyngeal samples by using a QIAmp Viral RNA Mini Kit (QIAGEN, Hamburg, Germany), according to manufacturer’s instructions. Reverse transcription was conducted with High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA) by using 20 μL of the previously extracted RNA. Respiratory viruses were identified by individual reverse transcription–PCRs or PCRs selective for RSV; influenza viruses A and B; human parainfluenza viruses 1, 2, and 3; human coronavirus; human metapneumovirus; adenovirus; human bocavirus; picornavirus; and the β-actin gene (9). For picornavirus, we used the primer pair OL26–OL27, which include a portion of the 5′ noncoding region (NCR) common to all picornaviruses, in the same conditions previously described (10). After sequence amplification, products were examined by capillary electrophoresis in an automated DNA sequencer (MegaBace, General Electric Healthcare–Amersham Biosciences, Little Chalfont, UK), and results were visualized through the MegaBace FragmentProfiler software, which discriminates fragment sizes and fluorescent intensities.
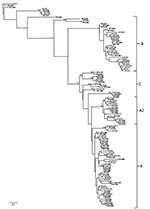
Samples in which picornavirus cDNA had been identified were submitted to a TaqMan-based real-time PCR protocol (11) to identify HRV and enterovirus. Only samples in which rhinovirus had been identified by real-time PCR were submitted to 5′ ΝCR sequencing with an ABI 377 automated sequencer (Applied Biosystems), and results were submitted to the GenBank database, accession nos. GU933027–GU933118. Sequencing of the 5′ NCR region has been shown to accurately discriminate among subtypes of rhinovirus, including genotypes C and A2 (12). All sequencing chromatograms obtained were edited manually to obtain contiguous fragments (contigs), by using Sequence Navigator software (Applied Biosystems). All sequences were screened at the National Center for Biotechnology Information website by using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). HRV genotype was confirmed by phylogenetic analysis as described below. Sequences generated and a set of reference strains representative of HRV genotypes, available in the GenBank database, were aligned by using ClustalW (www.ebi.ac.uk/Tools/clustalw2). Minor manual adjustments were made to improve the alignment with BioEdit software (www.mbio.ncsu.edu/BioEdit/BioEdit.html). The sequence of echovirus 11 was used as the outgroup. Phylogenetic analysis was performed with PAUP* version 4b10 (http://paup.csit.fsu.edu). Neighbor-joining and maximum-likelihood trees were constructed on the basis of appropriate nucleotide substitution models determined by Modeltest v3.7 (University of Vigo, Vigo, Spain). Bootstrapping was assessed by using 1,000 replicates. Trees were visualized by using the TreeView program (www.taxonomy.zoology.gla.ac.uk/rod/treeview.html).
To account for correlations among samples from the same patient, we used binomial generalized linear models to identify the virologic variables associated with the main endpoints (respiratory exacerbation and hospital admission). Results were presented as odds ratios (ORs) and 95% confidence intervals.
At least 1 respiratory virus was identified in 203 (49.8%) of 408 samples; rhinovirus was the main identified agent (139 samples, 34.1%). The results of virus identification, in relation to clinical status, are shown in Table 1. Co-infections were found in only 26 samples (6.4%); rhinovirus was the most frequent agent.
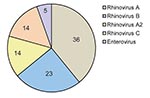
Sequencing was performed in 93 of 139 samples that were positive for rhinovirus and showed a predominance of genotype A (36 samples; 38.7%) (Figure 1). Rhinovirus subtypes A2 and C were identified in 14 samples each. Isolates in 3 samples were identified as HRV87, coxsackievirus, and echovirus. Therefore, 5 isolates were enteroviruses, which indicates that the TaqMan–based real-time PCR platform misidentified them. Sequencing was not successful in 1 sample. A maximum-likelihood phylogenetic tree of our samples and reference HRV sequences was constructed (Figure 2).
Patients were examined during acute exacerbation of respiratory disease on 142 occasions, and patients required hospital admission on 31 occasions. The identification of respiratory viruses was not associated with pulmonary exacerbations. Because rhinovirus was the main agent identified among stable patients, we verified the effects of respiratory viruses, excluding rhinovirus from the analysis. A significant association with respiratory exacerbation was found (OR 1.195, p = 0.010) (Table 2).
In contrast, when looking at rhinovirus subtypes, we noticed that identification of rhinovirus subtypes A2 or C was also significantly associated with respiratory exacerbations (OR 1.213) (Table 2). Identification of influenza A was the only variable associated with an increase in the risk for hospital admission (OR 1.988, 95% confidence interval 1.238–3.194, p = 0.004).
These new HRV genotypes were initially described in samples from patients with influenza-like illnesses (13). Recently, evidence has been increasing for the involvement of HRV-C in severe respiratory conditions such as bronchiolitis in infants (7) and in exacerbation of asthma (14). However, these studies that attempted to clarify the pathogenicity of these new species did not include control groups or nonsymptomatic persons.
In the study reported here, we obtained samples from patients during routine visits and exacerbations, which enabled us to identify a distinct role of different HRV subtypes. Our findings, however, cannot be extrapolated to infants with CF, which were underrepresented in our study. These infants may be at greater risk of HRV-C respiratory infections as they are for with RSV infections (2).
Previous studies of respiratory virus infections in CF patients provided conflicting results on the potential effect of rhinovirus. Smyth et al (15), who evaluated respiratory exacerbations in CF patients, identified rhinovirus as the leading agent, with exacerbations following a more benign course but resulting in greater use of intravenous antimicrobial drugs (15). Olesen et al. (4) studied 75 CF patients for 1 year and used PCR to investigate the presence of 7 different viruses in sputum or laryngeal aspirates. The group reported that viral infections did not reduce lung function or increase respiratory symptoms. Rhinovirus was by far the leading agent identified throughout their study (4). A more recent study by Wat et al. (3), in which they used real-time nucleic acid sequence–based amplification in conjunction with molecular markers to investigate the presence of 9 respiratory viruses in children with CF, described an association of viral infections with respiratory exacerbations, particularly those caused by influenza A, influenza B, and rhinovirus (3).
We report infections by the novel rhinovirus subtypes A2 and C in CF patients. A significant association was found between the presence of these agents and respiratory exacerbations. Our findings indicate the need for further investigation of HRV-C in CF patients.
Dr de Almeida is a pediatric pulmonologist at Instituto da Criança Hospital das Clínicas da Faculdade de Medicina de Universidade de São Paulo and a PhD candidate at the University of São Paulo. Her research interests are primarily related to respiratory diseases in children.
Acknowledgment
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo, grant 05/01625-8.
References
- Armstrong D, Grimwood K, Carlin JB, Carzino R, Hull J, Olinsky A, Severe viral respiratory infections in infants with cystic fibrosis. Pediatr Pulmonol. 1998;26:371–9. DOIPubMedGoogle Scholar
- Wat D, Gelder C, Hibbitts S, Cafferty F, Bowler I, Pierrepoint M, The role of respiratory viruses in cystic fibrosis. J Cyst Fibros. 2008;7:320–8. Epub 2008 Feb 6. DOIPubMedGoogle Scholar
- Olesen HV, Nielsen LP, Schiotz PO. Viral and atypical bacterial infections in the outpatient pediatric cystic fibrosis clinic. Pediatr Pulmonol. 2006;41:1197–204. DOIPubMedGoogle Scholar
- McErlean P, Shackelton LA, Andrews E, Webster DR, Lambert SB, Nissen MD, Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C). PLoS One. 2008; 3:e1847.
- Briese T, Renwick N, Venter M, Jarman RG, Ghosh D, Kondgen S, Global distribution of novel rhinovirus genotype. Emerg Infect Dis. 2008;14:944–7. DOIPubMedGoogle Scholar
- Lau SK, Yip CC, Tsoi HW, Lee RA, So LY, Lau YL, Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–64. DOIPubMedGoogle Scholar
- Wisdom A, Leitch EC, Gaunt E, Harvala H, Simmonds P. Screening respiratory samples for detection of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4–VP2 typing reveals high incidence and genetic diversity of HRV species C. J Clin Microbiol. 2009;47:3958–67. DOIPubMedGoogle Scholar
- da Silva Filho LVF, de Almeida MB, Flores TR, Tomikawa S, Wickbold D, Tateno AF, Clinical and functional impact of acute viral respiratory infections in cystic fibrosis. Pediatr Pulmonol. 2008;43:344.
- Papadopoulos NG, Sanderson G, Hunter J, Johnston SL. Rhinoviruses replicate effectively at lower airway temperatures. J Med Virol. 1999;58:100–4. DOIPubMedGoogle Scholar
- Deffernez C, Wunderli W, Thomas Y, Yerly S, Perrin L, Kaiser L. Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. J Clin Microbiol. 2004;42:3212–8. DOIPubMedGoogle Scholar
- Kiang D, Kalra I, Yagi S, Louie JK, Boushey H, Boothby J, Assay for 5′ noncoding region analysis of all human rhinovirus prototype strains. J Clin Microbiol. 2008;46:3736–45. DOIPubMedGoogle Scholar
- Lamson D, Renwick N, Kapoor V, Liu Z, Palacios G, Ju J, MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–402. DOIPubMedGoogle Scholar
- Khetsuriani N, Lu X, Teague WG, Kazerouni N, Anderson LJ, Erdman DD. Novel human rhinoviruses and exacerbation of asthma in children. Emerg Infect Dis. 2008;14:1793–6. DOIPubMedGoogle Scholar
- Smyth AR, Smyth RL, Tong CY, Hart CA, Heaf DP. Effect of respiratory virus infections including rhinovirus on clinical status in cystic fibrosis. Arch Dis Child. 1995;73:117–20. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 16, Number 6—June 2010
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Luiz Vicente Ferreira da Silva Filho, Rua Gregório Paes de Almeida, 1148 Vila Madalena, CEP 05450-001, São Paulo, SP, BrazilLuiz Vicente Ferreira da Silva Filho, Rua Gregório Paes de Almeida, 1148 Vila Madalena, CEP 05450-001, São Paulo, SP, Brazil
Top