Volume 17, Number 5—May 2011
Dispatch
Genetic Characterization of West Nile Virus Lineage 2, Greece, 2010
Figure
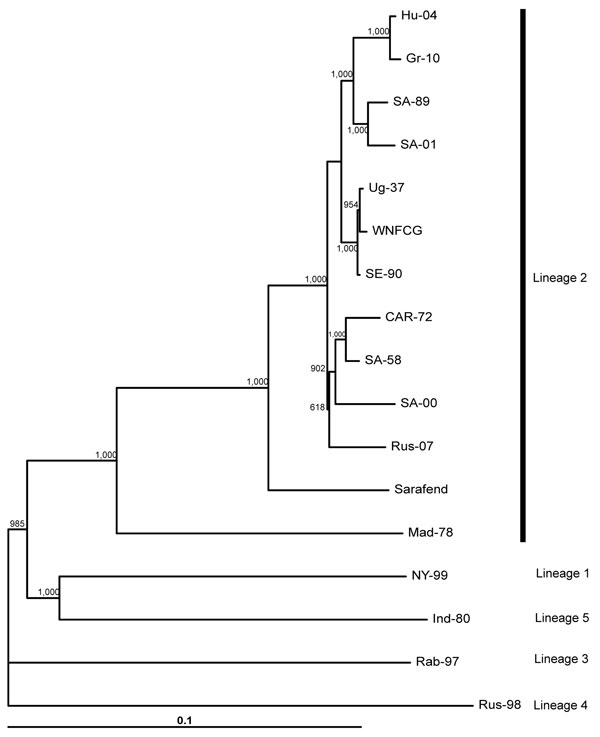
Figure. Neighbor-joining phylogram based on complete genome nucleotide sequences of selected West Nile virus strains. Strain abbreviations (isolation source, country, year, GenBank accession no.): Hu-04: Accipiter gentilis, Hungary, 2004, DQ116961; Gr-10: Culex pipiens, Greece, 2010, HQ537483; SA-89: human, South Africa, 1989, EF429197; SA-01: human, South Africa, 2001, EF429198; Ug-37: human, Uganda, 1937, AY532665; WNFCG: derivate of Ug-37, M12294; SE-90: Mimomyia lacustris, Senegal, 1990, DQ318019; CAR-72: Cx. tigripes, Central African Republic, 1972, DQ318020; SA-58: human, South Africa, 1958, EF429200; SA-00: human, South Africa, 2000, EF429199; Rus-07: human, Russia, 2007, FJ425721; Sarafend: derivate of Ug-37, AY688948; Mad-78: Coracopsis vasa, Madagascar, 1978, DQ176636; NY-99: human, USA, 1999, AF202541; Ind-80: human, India, 1980, DQ256476; Rab-97: Cx. pipiens, Czech Republic, 1997, AY765264; Rus-98: Dermacentor marginatus, Russia, 1998, AY277251. Rus-98 was used as outgroup. Bootstrap values of 1,000 replicates are shown. The main genetic lineages are indicated on the right. Horizontal bar shows genetic distance.