Human Parvovirus 4 in Nasal and Fecal Specimens from Children, Ghana
Jan Felix Drexler, Ulrike Reber, Doreen Muth, Petra Herzog, Augustina Annan, Fabian Ebach, Nimarko Sarpong, Samuel Acquah, Julia Adlkofer, Yaw Adu-Sarkodie, Marcus Panning, Egbert Tannich, Jürgen May, Christian Drosten, and Anna Maria Eis-Hübinger

Author affiliations: University of Bonn Medical Centre, Bonn, Germany (J.F. Drexler, U. Reber, D. Muth, A. Annan, F. Ebach, C. Drosten, A.M. Eis-Hübinger); Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany (P. Herzog, J. Adlkofer, E. Tannich, J. May); Kumasi Centre for Collaborative Research in Tropical Medicine, Kumasi, Ghana (A. Annan, N. Sarpong, S. Acquah); Kwame Nkrumah University of Science and Technology, Kumasi (Y. Adu-Sarkodie); and Freiburg University Medical Center, Freiburg, Germany (M. Panning)
Main Article
Figure 2
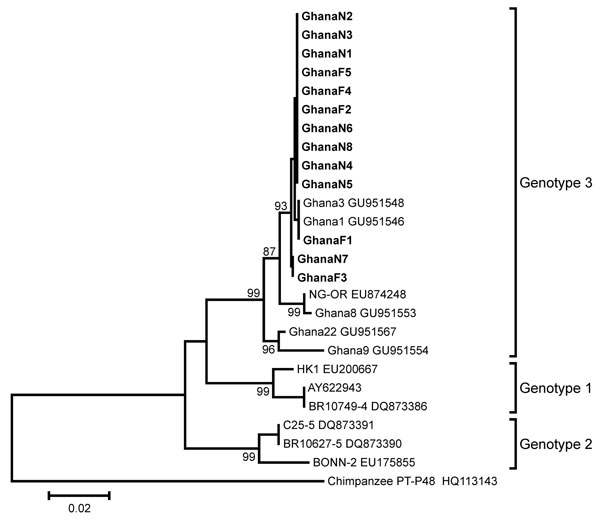
Figure 2. . . Phylogenetic analysis of a 483-nt fragment of the parvovirus 4 (PARV4) capsid-encoding open reading frame (ORF) 2 for PARV4 strains identified in children, Ghana. Neighbor-joining phylogeny was conducted in MEGA5.05 (www.megasoftware.net) by using a gap-free ORF2 fragment corresponding to positions 2,432–2,914 in the PARV4 genotype 3 prototype strain NG-OR (GenBank accession no. EU874248) with a nucleotide percentage distance substitution model and 1,000 bootstrap replicates. Scale bar indicates percentage uncorrected nucleotide distance. Previously published PARV4 sequences are given with strain names (if available) and GenBank accession numbers. Viruses newly identified are in boldface. The source of PARV4 strains identified in the study is indicated by capital letters (N, nasal specimen; F, fecal specimen). PARV4 genotypes are given to the right of taxa. A chimpanzee partetravirus was used as the outgroup.
Main Article
Page created: October 05, 2012
Page updated: October 05, 2012
Page reviewed: October 05, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
