Volume 18, Number 11—November 2012
Letter
Genome Sequencing of Pathogenic Rhodococcus spp.
Figure
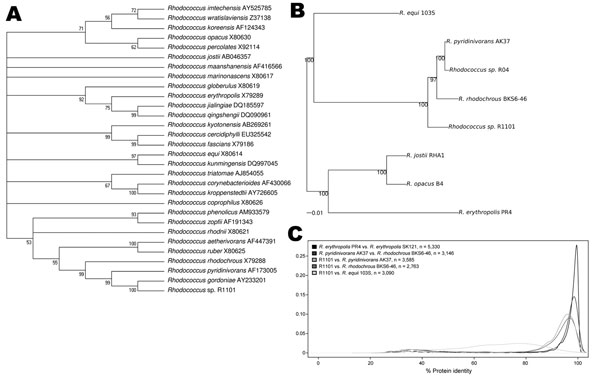
Figure. . . Phylogenetic and proteomic comparison of isolate R1101 and Rhodococcus spp. strains. A) 16S rRNA–based phylogenetic analysis. Complete or partial 16S rRNA gene sequences for 29 Rhodococcus spp. type strains were aligned with complete sequence of 16S rRNA gene for isolate R1101. GenBank accession numbers are provided. B) Genome-wide phylogenetic analysis. A sample of 67 protein-coding genes shared among genomes of isolate R1101 and 6 Rhodococcus spp. strains were analyzed. Scale bar indicates 0.01 nt substitutions per site. C) Genome-wide comparative proteome analysis. Density distributions of amino acid identity between homologous proteins from pairs of Rhodococcus strains are shown. The density distribution curves for R1101 show that R1101 is closely related, but not identical to, R. pyridinivorans (orange curve) and R. rhodochrous (red curve) at the protein level; R1101 is less closely related to R. pyridinivorans AK37 and R. rhodochrous than these 2 species are related to each other; and R1101 is most distantly related to R. equi (green curve).
1These authors contributed equally to this article.