Volume 18, Number 3—March 2012
Dispatch
Southeast Asian Foot-and-Mouth Disease Viruses in Eastern Asia
Cite This Article
Citation for Media
Abstract
Foot-and-mouth disease (FMD) outbreaks recently affected 2 countries (Japan and South Korea) in eastern Asia that were free of FMD without vaccination. Analysis of viral protein 1 nucleotide sequences indicated that FMD serotype A and O viruses that caused these outbreaks originated in mainland Southeast Asia to which these viruses are endemic.
Foot-and-mouth disease (FMD) is a highly contagious transboundary disease that affects domesticated animals and wildlife in Africa, Asia and parts of South America. Outbreaks of FMD in these disease-endemic regions continuously threaten livestock industries in countries that are free of FMD (with or without vaccination). The causative agent, FMD virus (FMDV), is a small, nonenveloped, picornavirus (genus Aphthovirus) that has 7 serotypes. This virus is easily transmitted by movement of infected livestock or animal products, contaminated persons, objects, and aerosols.
FMDV serotypes O, A, and Asia 1 are endemic to countries in mainland Southeast Asia (Cambodia, Laos, peninsula Malaysia, Myanmar, Thailand, and Vietnam) where regular outbreaks of FMD have been reported (1,2). Viral protein 1 (VP1) nucleotide sequence data are widely used for phylogenetic analyses (2,3) and have been used to characterize different FMDV lineages in Southeast Asia and track transboundary movements of the virus. Studies have shown close epidemiologic links between field outbreaks in countries in Southeast Asia (4–6).
During 2009–2010, the geographic range of 2 FMDV lineages endemic to Southeast Asia (serotypes A and O) expanded into eastern Asia and caused outbreaks in 6 countries in the region. Although outbreaks of FMD caused by serotype Asia 1 have been recently reported (2005–2009) (7), there have been no reported outbreaks caused by serotype A in eastern Asia since 1973.
Furthermore, serotype O strains from Southeast Asia have not been detected in countries in eastern Asia since 2004 when samples were sent to the Federal Centre for Animal Health (Vladimir, Russia) from Mongolia (GenBank accession no. JQ070317) (Figure A1). In 1999–2002, extensive outbreaks caused by the PanAsia strain of serotype O were reported in Japan, South Korea, China, Taiwan, and Russia (8–10). The purpose of this study was to determine the origins of recent FMD outbreaks in eastern Asia.
FMDVs characterized by antigen ELISA as serotype A were isolated from samples collected from FMD field outbreaks in Hubei Province, China, and in Gyeonggi-do Province, South Korea. Before FMD cases appeared in China in January 2009, serotype A infections had not occurred in that country China since 1964 (11), and until serotype A appeared in South Korea in January 2010, that country had been free of FMD without vaccination (for all FMDV serotypes) since 2002.
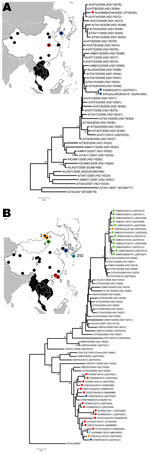
Phylogenetic analysis (12) showed that VP1 sequences obtained at Lanzhou Veterinary Research Institute (Gansu, China), the National Veterinary Research and Quarantine Service (Gyeonggi Do, South Korea), and the Institute of Animal Health (Pirbright, UK) were genetically similar to sequences obtained during 2008–2009 in Southeast Asia (Thailand and Malaysia), which belonged to the A/ASIA/Sea-97 lineage (Figure A1, panel A) (6). Additional FMD outbreaks caused by the same serotype that affected mainly cattle were reported in 6 provinces in China (Shanghai, Jiangsu, Guangxi, Guizhou, Shangdong, and Xinjiang) in 2009; in Beijing in January 2010 (Figure A1, panel A); and in Gyeonggi-do Province, South Korea, in January–March 2010 (6 cases).
Similar outbreaks occurred during 2010 that were caused by an FMD lineage of serotype O that is endemic to Southeast Asia (Figure A1, panel B). As of 2011, outbreaks caused by this serotype continue to occur across a wide region (Figure A1, panel B) and have affected China (including Hong Kong), South Korea, Japan, Mongolia, Russia, and North Korea.
Many FMD outbreaks (n = 292) were reported in Miyazaki Prefecture, Japan (April–June 2010). Fewer outbreaks (n = 18) were reported in China (in Guangdong, Gansu, Shaanxi, Jiangxi, and Guizhou Provinces; and in the autonomous regions of Ningxia, Xinjiang, Uyghur, and Tibet. Three outbreaks were reported in Hong Kong (February–March 2010); thirteen in South Korea (April 2010); nine, including 1 ongoing, in Mongolia (April 2010), and 2 in Russia (July and August 2010).
Japan had been free of FMD (without vaccination) since 2000, and Mongolia and Russia had not reported FMD outbreaks caused by serotype O since 2003 and 2004, respectively. These outbreaks have affected domesticated pigs, cattle, and small ruminants, and have spread to (gazelles, as represented by isolate O/MOG/9/2010) in Domod Province, Mongolia. A new increase in cases of FMD caused by serotype O has recently been reported in South Korea during December 2010 (>100 outbreaks) and in North Korea, and this serotype continues to pose a threat to livestock industries in the region.
VP1 sequences generated in the United Kingdom, China, Japan, South Korea, and Russia and those available in GenBank were analyzed by using by MEGA5 (12). Analysis showed that FMDVs causing serotype O outbreaks form 2 genetic clusters related to viruses within the Southeast Asia topotype (O/SEA/Mya-98 lineage), which are usually restricted to mainland Southeast Asia (Figure A1, panel B). There was >97.3% sequence relationship between sequences for FMDVs from China and those from outbreaks in Hong Kong, South Korea, and Japan.
Sequences for viruses collected in Mongolia were distinct (differing by 11.9% nt identity) and more closely related to other viruses collected during 2009 and 2010 in Southeast Asia (Thailand, Vietnam and Malaysia) (6). The 2 outbreaks in Russia (July and August 2010), which were caused by viruses from 2 of these sublineages, were located close to the borders with China and Mongolia and separated by ≈250 km. These outbreaks represent 2 distinct introductions of FMD into Russia.
Sequence data implicate regions of mainland Southeast Asia to which FMD is endemic as the source of serotype O and A FMDVs that have caused recent outbreaks in eastern Asia. These events are not unprecedented; a previous instance of spread of FMDV from Southeast Asia into China (Yunnan Province) in 2006 involved the Asia 1 serotype. Furthermore, FMDV O/SEA topotype (Mya-98) was also detected in China in 2003 and in Mongolia in 2004. These findings provide evidence for the porous nature of borders between mainland Southeast Asia and neighboring countries and highlight the continued threat posed by FMD as a transboundary disease in the region. The extent to which viruses have spread into countries that were previously free of FMD (without vaccination) is a cause for concern.
Although VP1 sequence data can be used to characterize the viruses that are causing these outbreaks, further coordination and sharing of sequence data are now urgently required to formally identify transboundary transmission links between affected countries in the region. Complete FMDV genome sequence analyses from these field cases and additional material may provide a suitable approach to reconstruct high-resolution transmission trees and connect clusters of outbreaks (13,14).
This report describes recent incursion of FMDVs from Southeast Asia into eastern Asia. In vitro vaccine matching data (from the Institue of Animal Health) indicate that currently available vaccine strains (A/May/97and O/Manisa) should protect against representative isolates of these 2 serotypes. However, close monitoring of antigenicity and of the spread of these lineages from Southeast Asia is essential to ensure that risks for further and continued outbreaks can be mitigated.
Mr Knowles is a molecular virologist at the Institute for Animal Health in Pirbright, UK. His research interests are the molecular epidemiology and evolution of picornaviruses of animals, particularly foot-and-mouth disease virus.
Acknowledgments
We thank researchers in Southeast Asia for providing data and samples, and Wilai Linchongsubongkoch and colleagues at the Regional Reference Laboratory in Pakchong, Thailand for assistance. Nucleotide alignments used for phylogenetic analyses are available by request from the corresponding author.
This study was supported by the Department for Environment, Food and Rural Affairs (SE2939), the Food and Agriculture Organization (PR 41764), and the Network of Excellence for Epizootic Disease Diagnosis and Control IC4.7 projects (contract no. FOOD-CT-2006-016236).
References
- Gleeson LJ. A review of the status of foot-and-mouth disease in South-East Asia and approaches to control and eradication. Rev Sci Tech. 2002;21:465–75.PubMedGoogle Scholar
- Rweyemamu M, Roeder P, Mackay D, Sumption K, Brownlie J, Leforban Y, Epidemiological pattern of foot-and-mouth disease worldwide. Transbound Emerg Dis. 2008;55:57–72. DOIPubMedGoogle Scholar
- Knowles NJ, Samuel AR. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 2003;91:65–80. DOIPubMedGoogle Scholar
- Le VP, Nguyen T, Lee K-N, Ko Y-J, Lee H-S, Nguyen VC, Molecular characterization of serotype A foot-and-mouth disease virus circulating in Vietnam in 2009. Vet Microbiol. 2010;144:58–66. DOIPubMedGoogle Scholar
- Le VP, Nguyen T, Park J-H, Kim S-M, Ko Y-J, Lee H-S, Heterogeneity and genetic variations of serotypes O and Asia 1 foot and mouth disease viruses isolted in Vietnam. Vet Microbiol. 2010;145:220–9. DOIPubMedGoogle Scholar
- Abdul-Hamid NF, Hussein NM, Wadsworth J, Radford AD, Knowles NJ, King DP. Phylogeography of foot-and-mouth disease virus types O and A in Malaysia and surrounding countries. Infect Genet Evol. 2011;11:320–8. DOIPubMedGoogle Scholar
- Valarcher J-F, Knowles NJ, Zakharov V, Scherbakov A, Zhang Z, Shang Y-J, Multiple origins of foot-and-mouth virus serotype Asia 1 outbreaks, 2003–2007. Emerg Infect Dis. 2009;15:1046–51. DOIPubMedGoogle Scholar
- Knowles NJ, Samuel AR, Davies PR, Midgley RJ, Valarcher JF. Pandemic strains of foot-and-mouth disease virus serotype O. Emerg Infect Dis. 2005;11:1887–93.PubMedGoogle Scholar
- Sakamoto K, Kanno T, Yamakawa M, Yoshida K, Yamazoe R, Murakami Y. Isolation of foot-and-mouth disease virus from Japanese black cattle in Miyazaki Prefecture, Japan 2000. J Vet Med Sci. 2002;64:91–4. DOIPubMedGoogle Scholar
- Wee S-H, Park J-Y, Joo Y-S, Lee J-H, An S-H. Control measures implemented during the 2002 foot-and-mouth disease outbreak in the Republic of Korea. Vet Rec. 2004;154:598–600. DOIPubMedGoogle Scholar
- Liu Z, Zhao Q, Liu W, Zhou P, Zhu C, Chang H, Analysis of VP1-coding nucleotide sequences of six strains of foot-and-mouth disease virus type A. Chin J Virol. 1998;14:60–7.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony Methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Cottam EM, Haydon DT, Paton DJ, Gloster J, Wilesmith JW, Ferris NP, Molecular epidemiology of foot-and-mouth disease virus outbreak in the United Kingdom. J Virol. 2006;80:11274–82. DOIPubMedGoogle Scholar
- Cottam EM, Wadsworth J, Shaw AE, Rowlands RJ, Goatley L, Maan S, Transmission pathways of foot-and-mouth disease virus in the United Kingdom in 2007. PLoS Pathog. 2008;4:e1000050. DOIPubMedGoogle Scholar
Figure
Cite This ArticleTable of Contents – Volume 18, Number 3—March 2012
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Donald P. King, Institute for Animal Health, Ash Rd, Pirbright, Surrey, GU24 0NF, UK
Top