Novel Orthobunyavirus in Cattle, Europe, 2011
Bernd Hoffmann
1, Matthias Scheuch
1, Dirk Höper, Ralf Jungblut, Mark Holsteg, Horst Schirrmeier, Michael Eschbaumer, Katja V. Goller, Kerstin Wernike, Melina Fischer, Angele Breithaupt, Thomas C. Mettenleiter, and Martin Beer

Author affiliations: Friedrich-Loeffler-Institut, Greifswald–Insel Riems, Germany (B. Hoffmann, M. Scheuch, D. Höper, H. Schirrmeier, M. Eschbaumer, K.V. Goller, K. Wernike, M. Fischer, A. Breithaupt, T.C. Mettenleiter, M. Beer); State Veterinary Diagnostic Laboratory, Arnsberg, Germany (R. Jungblut); Chamber of Agriculture for North Rhine-Westphalia, Bovine Health Service, Bonn, Germany (M. Holsteg)
Main Article
Figure 2
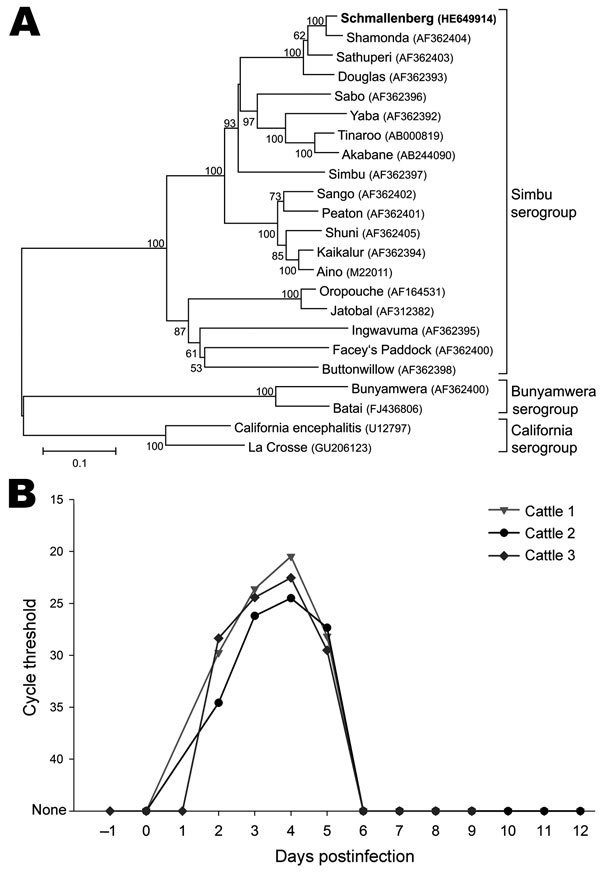
Figure 2. A) Phylogenetic relationship between Schmallenberg virus and orthobunyaviruses of the Simbu, Bunyamwera, and California serogroups. International Nucleotide Sequence Database Collaboration accession numbers of the sequences in the analysis are indicated in the tree. The neighbor-joining tree is based on the nucleocapsid gene of the small segment (702 nt). Numbers at nodes represent the percentage of 1,000 bootstrap replicates (values <50 are not shown). Scale bar indicates the estimated number of nt substitutions per site. B) Detection of Schmallenberg virus genome in the blood of experimentally infected calves. The highest genome copy number was detected on postinoculation day 4.
Main Article
Page created: February 16, 2012
Page updated: February 16, 2012
Page reviewed: February 16, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
