Volume 18, Number 7—July 2012
Dispatch
Multiple Introductions of Avian Influenza Viruses (H5N1), Laos, 2009–2010
Abstract
Avian influenza viruses (H5N1) of clades 2.3.4.1, 2.3.4.2, and 2.3.2.1 were introduced into Laos in 2009–2010. To investigate these viruses, we conducted active surveillance of poultry during March 2010. We detected viruses throughout Laos, including several interclade reassortants and 2 subgroups of clade 2.3.4, one of which caused an outbreak in May 2010.
Since 2003, highly pathogenic avian influenza virus (H5N1) has spread from southern China throughout Southeast Asia and to Europe and Africa (1,2). Since 2003, Laos has experienced outbreaks of clade 1 (2003), clade 2.3.4 (2006, 2007, 2008, 2009, 2010), and clade 2.3.2 viruses (twice in 2008) (3,4). Active surveillance of domestic ducks and chickens in Laos has been limited, but serum antibodies against subtypes H5 and H9 have been detected in ducks. In addition, subtype H5N1 virus was isolated from healthy ducks in 2006, and subtype H3N8 virus was detected in 2007 (4,5). To explore the diversity, extent, and endemicity of avian influenza viruses in Laos, we conducted a survey of healthy domestic poultry throughout the country in March 2010.
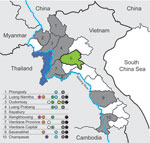
Serum samples were collected in 9 of 17 provinces in Laos from healthy ducks and chickens in live-bird markets, village backyard flocks, and layer duck farms. Cloacal, tracheal, and environmental (fecal and water) swab specimens were also collected and placed immediately in transport medium (Figure). Swab specimens were screened in pools of 4 by using a real-time reverse transcription PCR for the matrix (M) gene segment (6). Positive pools were reextracted individually, retested, tested for hemagglutinin 5 (H5) by real-time reverse transcription PCR (7), and injected into 10–11-day-old embryonated chicken eggs.
Sequencing was conducted by using an Illumina (San Diego, CA, USA) platform (swabs and isolates) (8,9) and conventional Sanger sequencing (isolates). Illumina reads were first mapped to a database of publicly available human, avian, and swine influenza virus reference sequences from the Western Hemisphere Americas and Eurasia, and then remapped against references with the highest number of reads and best average coverage. Final average coverage varied between samples and segments with ranges of 123–24,163 (9 samples), 21–18,671 (12 samples) and 9–436 for sample A/duck/Lao/670/10. Isolate genotypes were verified by using Sanger sequencing. Mixed infections could not be excluded for direct sequencing in the absence of an isolate.
Phylogenetic analysis (ClustalW [http://www.clustal.org/], neighbor-joining analysis, 1,000 bootstrap tests, maximum composite likelihood, pairwise deletions) was conducted by using MEGA version 5.02 (10). Sequences are available in GenBank (CY098294–CY098334, CY098336–CY098340, CY098342–CY098368, and CY098370–CY098464). Serum samples were screened by using an ELISA (FlockChek MultiS Screen; IDEXX Laboratories, Westbrook, ME, USA), and antibody-positive serum samples were tested by using a hemagglutinin inhibition assay for subtypes H3, H4, H5 (clades 2.3.2 and 2.3.4), H6, and H9 (lineages G1 and Y280) as described (11).
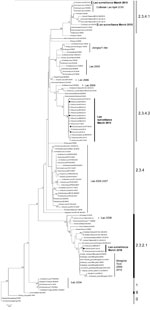
During March 2010, a total of 3,695 swab specimens were collected (1,928 duck and 279 chicken cloacal samples, 446 duck tracheal samples, 675 fecal samples, and 367 water samples). M gene prevalence was 4.0% (ducks), 1.8% (chickens), and 0.3% (environment samples). Five isolates were obtained (Figure A1). All M gene–positive swab specimens were collected in 13 locations (8 backyards, 2 markets, and 3 farms) (Table 1; Figure) Sample protection was suboptimal, and only 21 samples could be subtyped by real-time RT-PCR and sequencing (Table 1).
Phylogenetic analysis identified 3 groups of viruses: clades 2.3.2.1, 2.3.4.1, and 2.3.4.2 (Figure A1). Two samples were closely related to A/chicken/Lao/LH1/2010–like virus (outbreak in Vientiane in April–May 2010) and to A/chicken/Laos/C100209–194-PTK/2009–like virus (outbreak in Phongsaly in February 2009) (clade 2.3.4.1) (Figure A1; Table 1). These viruses were closely related to A/Guizhou/1/2009 and A/chicken/Vietnam/NCVD-404/2010 (Figure A1).
Sixteen surveillance viruses of clade 2.3.4.2 were highly homogeneous (Figure A1; Table 1) and closely related to A/environment/Guizhou/4/2009 and A/chicken/Vietnam/NCVD-394/2010–like viruses, although they contained the polymerase basic 2 (PB2) gene of a clade 2.3.2.1 donor virus and were therefore interclade reassortants (Figure A1; Table 1). The genotype of the 4 isolates was verified by using Sanger sequencing. The PB2 gene was most closely related to A/grey_heron/Hong_Kong/1046/2008–like viruses. These reassortants were detected in 3 locations in northern (backyard), central (backyard), and southern (farm) Laos, suggesting multiple introductions of reassortants into Laos (Figure A1; Table 1).
Three clade 2.3.2.1 viruses were detected in 2 ducks and 1 environmental sample (same trader). These viruses were interclade reassortants. Two (1 isolate and 1 direct sequence) contained 6 or 7 segments that were A/whooper swan/Mongolia/6/2009 like (clade 2.3.2.1), and the nucleoprotein gene was A/tree_sparrow/Jiangsu/1/08 like (clade 2.3.4). The environmental sample (direct sequence) contained A/whooper swan/Mongolia/6/2009–like hemagglutinin and M genes and A/Guizhou/1/2009–like PB2, nucleoprotein, and neuraminidase genes (clade 2.3.4.1) (Table 1). The genotype of the isolate was verified by using Sanger sequencing. Hemagglutinin segments of the reassortants were identical (100% nt identity). This identity and the source of the 3 samples (1 trader) suggest that reassortment occurred recently, likely in Laos.
Antibody titers to H5 and H9 and a subtype H3N8 virus isolate have been reported in Laos (4,5). Subtypes H4 and H6 also circulate in this region (1,2,12). For this study, 2,148 serum samples (1,899 from ducks, 200 from chickens, and 49 from unspecified species) were collected and 267 antibody-positive (seroprevalence 14%) duck and 15 (7.5%) chicken serum samples were detected. Hemagglutination inhibition testing for specific antibodies against H3, H4, H5, H6, and H9 detected all antibodies but to H3 (Table 2). Antibodies against H9 were detected at highest titers and most frequently (1.1% of ducks for each H9 lineage), although some cross-reactivity between G1 and Y280 likely occurred (Table 2). These ducks were most widely distributed (18 [19%] of 97 locations in all 9 provinces). Antibodies against H5 clade 2.3.4 were found at the detection limit in few serum samples (0.4% of ducks in 3 northern and 2 southern provinces). Antibodies against H5 clade 2.3.2 were detected in 0.4% of ducks in 4 northern provinces and the capital of Vientiane.
Among ducks, 34 (1.7%) were either shedding or had antibodies against avian influenza virus (H5N1), and there was >1 virus-positive or antibody-positive duck in each of the 9 provinces sampled. One village had 36% of sampled ducks exposed to this virus; 18% shed 2.3.4 virus and 18% had antibody to 2.3.2 virus (Table 2). One duck in this village had antibodies against 2.3.2 and 2.3.4 clade viruses. Exposure to 2.3.2 and 2.3.4 viruses was evident in ducks from locations in 3 other provinces (1 district each in Champasak and Xiengkhoung, and several districts in Luang Prabang).
This study showed that 3 groups of avian influenza viruses (H5N1) were likely introduced into Laos in 2009–2010, one of which resulted in 2 outbreaks (2009, 2010). In all 9 provinces where surveillance was conducted, ducks had been exposed to this virus. Evidence of clades 2.3.2 and 2.3.4 virus activity was detected in 4 provinces. Several interclade reassortants were identified, demonstrating the high genetic mobility of these viruses in the region. Since 2004, Laos has had repeated outbreaks of highly pathogenic avian influenza viruses, which have also been detected in China and Vietnam. There is no evidence that a particular virus lineage has established itself in Laos. The frequency of introduction, diversity, and extent of these viruses in Laos suggests considerable movement of viruses into the country from surrounding territories (China and Vietnam, but not Cambodia) and within the country.
Dr Sonnberg is a postdoctoral research associate at St. Jude Children’s Research Hospital, Memphis, Tennessee. Her research interests are surveillance, transmission propensity, and reassortment of influenza viruses.
Acknowledgments
We thank Jennifer DeBeauchamp, Scott Krauss, Mariette Ducatez, David Walker, Jerry Parker, Richard Elia, Betsy Williford, and David Galloway for technical assistance, data management, and help in preparing the figures and manuscript; and the Australian Animal Health Laboratory (Geelong, Victoria, Australia) for provision of Lao H5 sequences of avian influenza virus.
This study was supported by US Agency International Development project OSRO/RAS/604/USA Baby 3, National Institutes of Health contract HHSN266200700005C, and American Lebanese Syrian Associated Charities.
References
- Alexander DJ. Summary of avian influenza activity in Europe, Asia, Africa, and Australasia, 2002–2006. Avian Dis. 2007;51(Suppl):161–6. DOIPubMedGoogle Scholar
- Brown IH. Summary of avian influenza activity in Europe, Asia, and Africa, 2006–2009. Avian Dis. 2010;54(Suppl):187–93. DOIPubMedGoogle Scholar
- World Organisation for Animal Health. Update on highly pathogenic avian influenza in animals (type H5 and H7) [cited 2012 May 1]. http://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/2011/
- Boltz DA, Douangngeun B, Phommachanh P, Sinthasak S, Mondry R, Obert C, Emergence of H5N1 avian influenza viruses with reduced sensitivity to neuraminidase inhibitors and novel reassortants in Lao People’s Democratic Republic. J Gen Virol. 2010;91:949–59. DOIPubMedGoogle Scholar
- Boltz DA, Douangngeun B, Sinthasak S, Phommachanh P, Rolston S, Chen H, H5N1 influenza viruses in Lao People’s Democratic Republic. Emerg Infect Dis. 2006;12:1593–5. DOIPubMedGoogle Scholar
- World Health Organization. CDC protocol of real-time RT-PCR for swine influenza A (H1N1) [cited 2012 May 1]. http://www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCRprotocol_20090428.pdf
- Centers for Disease Control and Prevention (CDC) CDC Realtime RT-PCR. (rRTPCR) protocol for detection and characterization of influenza (version 2007). CDC ref. no. I-007–05. Atlanta: The Centers; 2007.
- Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A viruses. J Virol. 2009;83:10309–13. DOIPubMedGoogle Scholar
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–89. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Palmer DF, Dowdle WR, Coleman MT, Schild GC. Advanced laboratory techniques for influenza diagnosis. Immunology Series no. 6. Atlanta: Center for Disease Control; 1975.
- Global Initiative on Sharing All Influenza Data. EpiFlu [cited 2012 May 1]. http://platform.gisaid.org
Figures
Tables
Cite This ArticleTable of Contents – Volume 18, Number 7—July 2012
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Stephanie Sonnberg, Division of Virology, St. Jude Children’s Research Hospital, MS 330, Memphis, TN 38105, USA
Top