Volume 18, Number 7—July 2012
Dispatch
Multiple Introductions of Avian Influenza Viruses (H5N1), Laos, 2009–2010
Figure A1
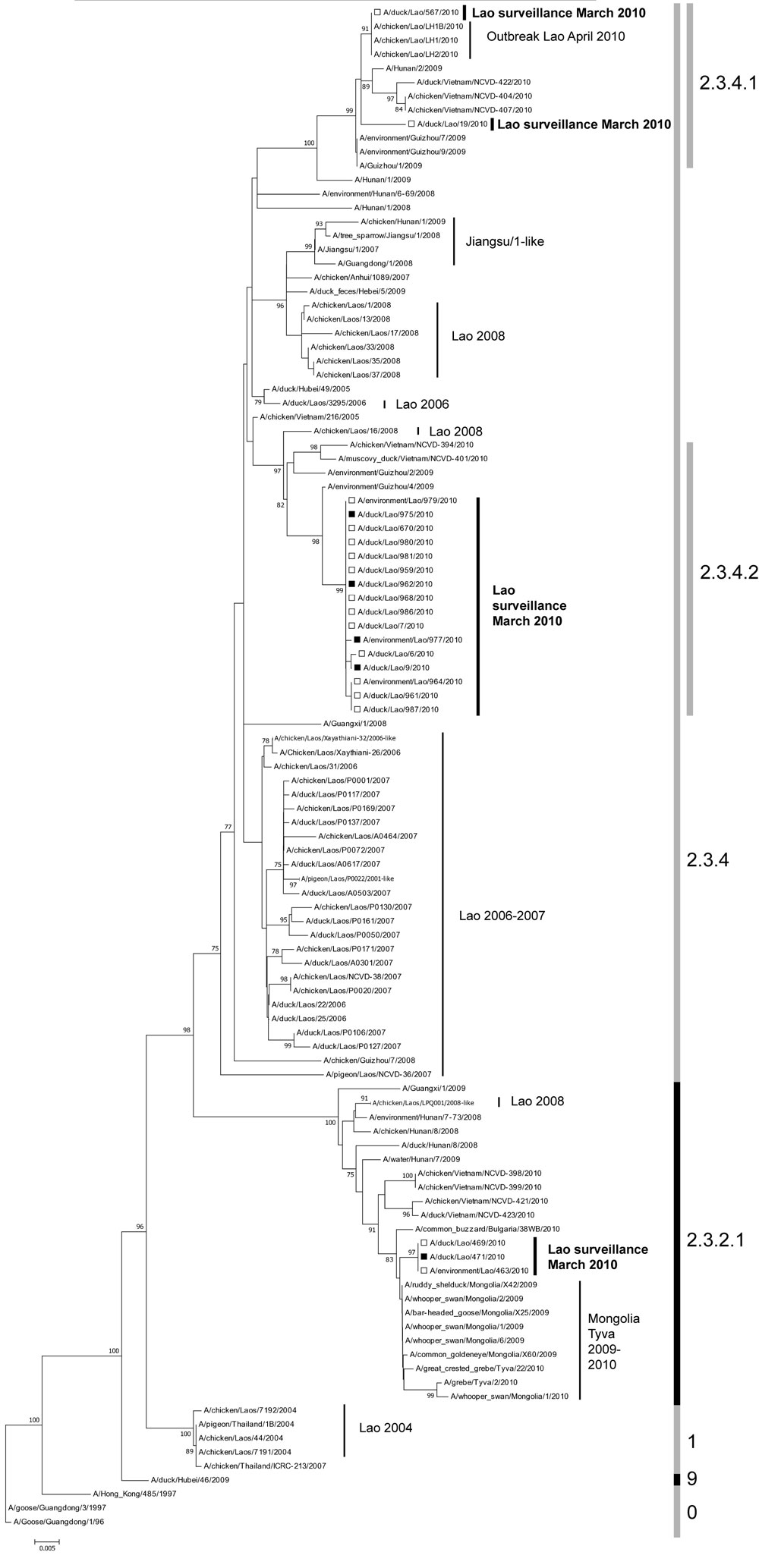
Figure A1. . Phylogenetic relationships of hemagglutinin 5 (H5) gene segments of avian influenza viruses (H5N1), Laos, 2009–2010. The tree was rooted in A/goose/Guangdong/1/1996 (nt 91–919, H5 numbering). Surveillance sequences are indicated in boldface and marked with black (isolates) or open (direct sequences) squares. Bootstrap values >75 are shown. Clade numbers are shown on the right. Scale bar indicates nucleotide substitutions per site. A/chicken/Laos/Xayathiani-32/2006-like, A/pigeon/Laos/P0022/2007-like, and A/chicken/Laos/LPQ001/2008-like viruses each represent 3 sequences with identical hemagglutinins. Reference viruses were obtained through EpiFlu of the Global Initiative on Sharing All Influenza Data and provided by the following laboratories in addition to sequences already in the public domain: A/chicken/Vietnam/NCVD-407/2010, A/chicken/Vietnam/NCVD-394/2010, A/chicken/Vietnam/NCVD-398/2010, A/chicken/Vietnam/NCVD-399/2010, A/duck/Vietnam/NCVD-422/2010, A/chicken/NCVD-404/2010, A/Muscovy_duck/Vietnam/NCVD-401/2010, A/chicken/Vietnam/NCVD-421/2010 A/duck/Vietnam/NCVD-423/2010 (original laboratory: National Centre of Veterinary Diagnostics, Hanoi, Vietnam; submitting laboratory: Centers for Disease Control and Prevention, Atlanta, GA, USA); A/Hunan/2/2009, A/Hunan/1/2009, A/Guizhou/1/2009, A/Guangxi/1/2009, A/environment/Guizhou/2/2009, A/environment/Guizhou/4/2009, A/environment/Guizhou/7/2009, A/environment/Guizhou/9/2009, A/water/Hunan/7/2009A/Guangxi/1/2008, A/Guangdong/1/2008, A/Hunan/1/2008, A/duck feces/Hebei/5/2009 (original and submitting laboratory: World Health Organization Chinese National Influenza Center, Beijing, China), Jiangsu/1-like–A/tree_sparrow/Jiangsu/1/2008–like viruses.