Multiple Synchronous Outbreaks of Puumala Virus, Germany, 2010
Jakob Ettinger
1, Jorg Hofmann
1, Martin Enders, Friedemann Tewald, Rainer M. Oehme, Ulrike M. Rosenfeld, Hanan Sheikh Ali, Mathias Schlegel, Sandra Essbauer, Anja Osterberg, Jens Jacob, Daniela Reil, Boris Klempa, Rainer G. Ulrich, and Detlev H. Kruger

Author affiliations: Charité Medical School and Labor Berlin Charité-Vivantes GmbH, Berlin, Germany (J. Ettinger, J. Hofmann, B. Klempa, D.H. Kruger); Institute of Virology, Infectious Diseases, and Epidemiology, Stuttgart, Germany (M. Enders, F. Tewald); Baden-Wuerttemberg State Health Office, Stuttgart (R.M. Oehme); Friedrich-Loeffler Institut, Greifswald, Germany (U.M. Rosenfeld, H.S. Ali, M. Schlegel, R.G. Ulrich); Bundeswehr Institute of Microbiology, Munich, Germany (S. Essbauer, A. Osterberg); Institute for Plant Protection in Horticulture and Forestry, Münster, Germany (J. Jacob, D. Reil); and Slovak Academy of Sciences, Bratislava, Slovakia (B. Klempa)
Main Article
Figure 1
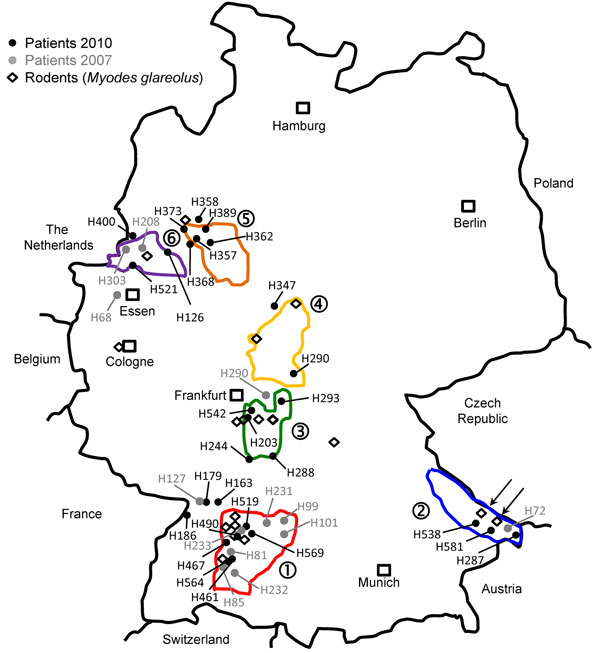
Figure 1. .
. . Distribution of investigated Puumala virus infections in Germany. Black dots indicate sequences obtained from patient samples in 2010; gray dots indicate sequences obtained from patient samples in 2007; diamonds indicate sequences obtained from rodent (Myodes glareolus) samples. Areas surrounded by lines indicate outbreak regions (numbered 1–6) where Puumala virus nucleotide sequences of human and vole origin have been analyzed. Numbers of the outbreak regions/virus clades and designations of local virus strains are also used in Figure 2. Arrows in outbreak region no. 2 indicate trapping sites of rodents from which strains 10 MuEb14v and 10 MuEb51 were obtained, which originated from localities 25 km apart.
Main Article
Page created: August 16, 2012
Page updated: September 05, 2012
Page reviewed: September 05, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
