Volume 18, Number 9—September 2012
Letter
Picobirnaviruses in the Human Respiratory Tract
Figure
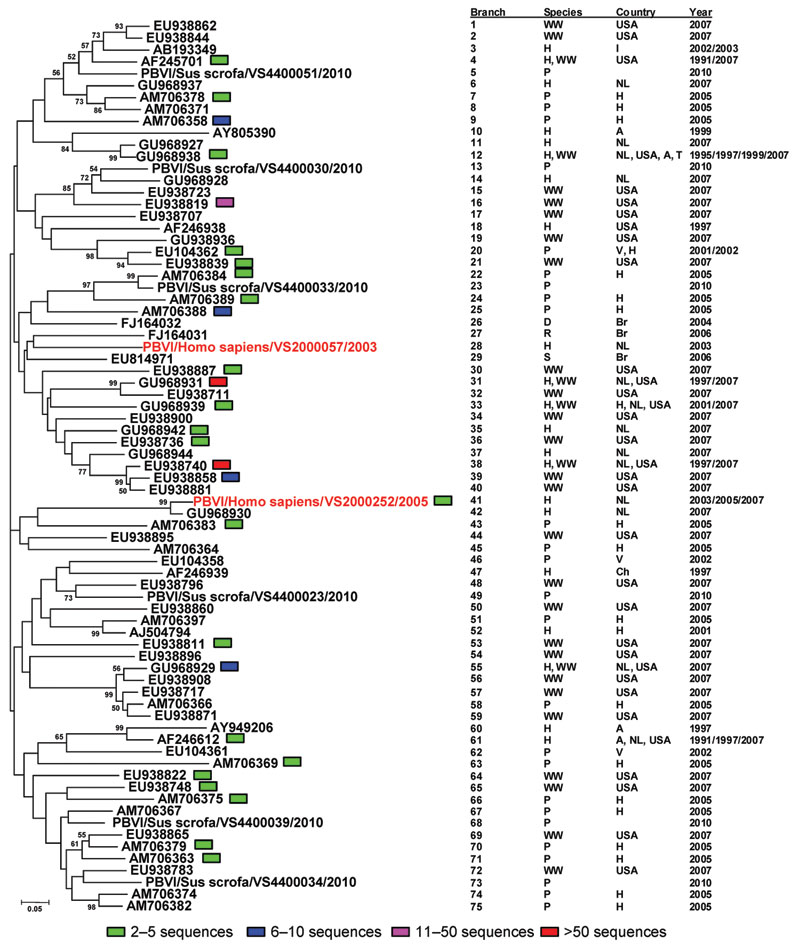
Figure. . Neighbor-joining (Jukes-Cantor model) phylogenetic tree of an ≈165-bp fragment of the genogroup I picobirnavirus RNA-dependent RNA polymerase gene from known human, porcine, and wastewater genogroup I picobirnaviruses and newly characterized genogroup I picobirnaviruses (sequences are available on request) from the human respiratory tract. Each branch represents a sequence or group of sequences (95% identical with gaps) indicated by the presence of a colored block. Every branch corresponds to the adjacent table, which explains the source, year of isolation, and location of the virus variants. Reference sequences from previously published studies are identified by their GenBank accession numbers. The designations for picobirnaviruses from various species are designated as follows: P, porcine; WW, wastewater; D, dog; S, snake; R, rat; H, human; The different countries of isolation are designated as follows: A, Argentina; Br, Brazil; Ch, China; H, Hungary; I, India; NL, the Netherlands; USA, United States of America; T, Thailand; V, Venezuela. The sequences from the newly identified human respiratory picobirnaviruses in this study comply with recent nomenclature proposals (9) and are indicated in red.
References
- Bányai K, Jakab F, Reuter G, Bene J, Uj M, Melegh B, Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch Virol. 2003;148:2281–91. DOIPubMedGoogle Scholar
- Bányai K, Martella V, Bogdan A, Forgach P, Jakab F, Meleg E, Genogroup I picobirnaviruses in pigs: evidence for genetic diversity and relatedness to human strains. J Gen Virol. 2008;89:534–9. DOIPubMedGoogle Scholar
- Bhattacharya R, Sahoo GC, Nayak MK, Rajendran K, Dutta P, Mitra U, Detection of genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect Genet Evol. 2007;7:229–38. DOIPubMedGoogle Scholar
- Rosen BI, Fang ZY, Glass RI, Monroe SS. Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology. 2000;277:316–29. DOIPubMedGoogle Scholar
- Giordano MO, Martinez LC, Rinaldi D, Guinard S, Naretto E, Casero R, Detection of picobirnavirus in HIV-infected patients with diarrhea in Argentina. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18:380–3. DOIPubMedGoogle Scholar
- Martínez LC, Giordano MO, Isa MB, Alvarado LF, Pavan JV, Rinaldi D, Molecular diversity of partial-length genomic segment 2 of human picobirnavirus. Intervirology. 2003;46:207–13. DOIPubMedGoogle Scholar
- Smits SL, Poon LL, van LM, Lau PN, Perera HK, Peiris JS, et al. Genogroup I and II picobirnaviruses in respiratory tracts of pigs. Emerg Infect Dis. 2011;17:2328–30. DOIPubMedGoogle Scholar
- van Leeuwen M, Williams MM, Koraka P, Simon JH, Smits SL, Osterhaus AD. Human picobirnaviruses identified by molecular screening of diarrhea samples. J Clin Microbiol. 2010;48:1787–94. DOIPubMedGoogle Scholar
- Fregolente MC, Gatti MS. Nomenclature proposal for picobirnavirus. Arch Virol. 2009;154:1953–4. DOIPubMedGoogle Scholar
- Yu Y, Breitbart M, McNairnie P, Rohwer F. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics. 2006;7:57. DOIPubMedGoogle Scholar