Volume 19, Number 1—January 2013
Research
Novel Polyomavirus associated with Brain Tumors in Free-Ranging Raccoons, Western United States
Figure 3
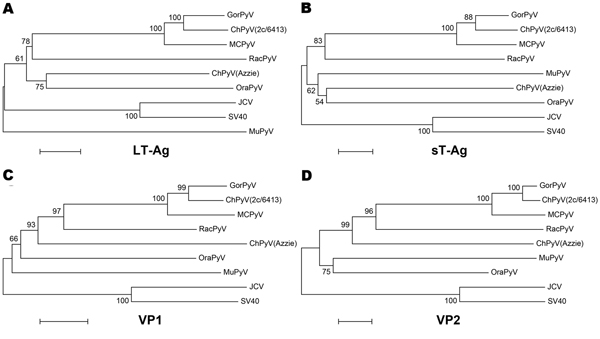
Figure 3. . Phylogenetic relationship of RacPyV with representative polyomavirus species. Phylogenetic trees were individually generated on the basis of amino acid sequences of LT-Ag (A), sT-Ag (B), VP1, and VP2 (C, D) by using the neighbor-joining method with p-distance and 1,000 bootstrap replications. RacPyV, raccoon polyomavirus; LT-Ag, large T-antigen; sT-Ag, small T-antigen; VP, viral protein; GorPyV, gorilla polyomavirus; ChPyV, chimpanzee polymavirus; MCPyV, Merkel cell polyomavirus; OraPyV, orangutan polyomavirus; JCV, JC virus; SV40, simian virus 40; MuPyV, murine polyomavirus. The bar represents 5% estimated phylogenetic divergence.
Page created: December 21, 2012
Page updated: December 21, 2012
Page reviewed: December 21, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.