Volume 19, Number 12—December 2013
Dispatch
Novel Variants of Clade 2.3.4 Highly Pathogenic Avian Influenza A(H5N1) Viruses, China
Figure
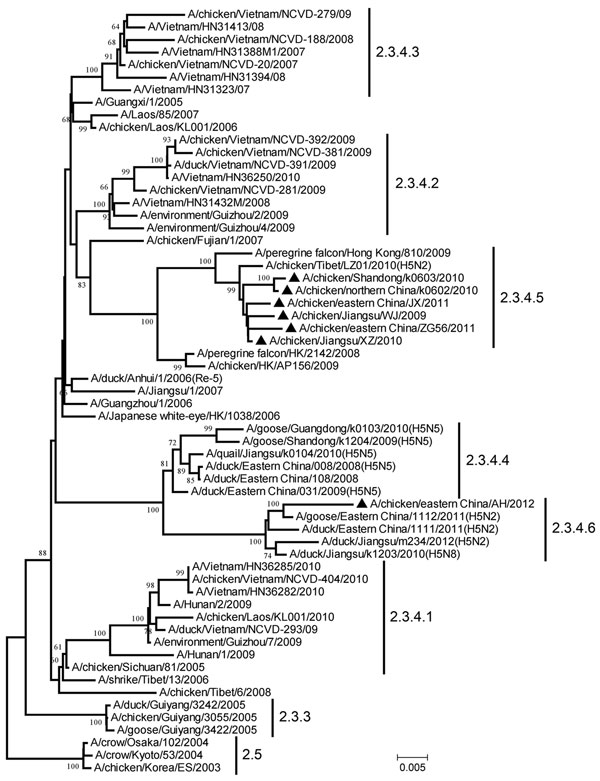
Figure. . Phylogenetic tree of the hemagglutinin (HA) genes of the diverged avian influenza H5 subtype clade 2.3.4 variants from China and reference sequences retrieved from the GenBank database and partially recommended by the World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization of the United Nations H5N1 Evolution Working Group The neighbor-joining tree was generated by using MEGA 5.1 software (www.megasoftware.net). Numbers above or below the branch nodes denote bootstrap values >60% with 1,000 replicates. Numbers on the right are existing (2.3.3, 2.3.4.1, 2.3.4.2, 2.3.4.3, 2.5) and proposed (2.3.4.4, 2.3.4.5, 2.3.4.6) virus subclades. Black triangles indicate the 7 variants identified in this study; GenBank accession numbers for their HA genes are KC631941–KC631946 and KC261450. Scale bar indicates nucleotide substitutions per site.