Volume 19, Number 4—April 2013
Letter
Hand, Foot and Mouth Disease Outbreak and Coxsackievirus A6, Northern Spain, 2011
Figure
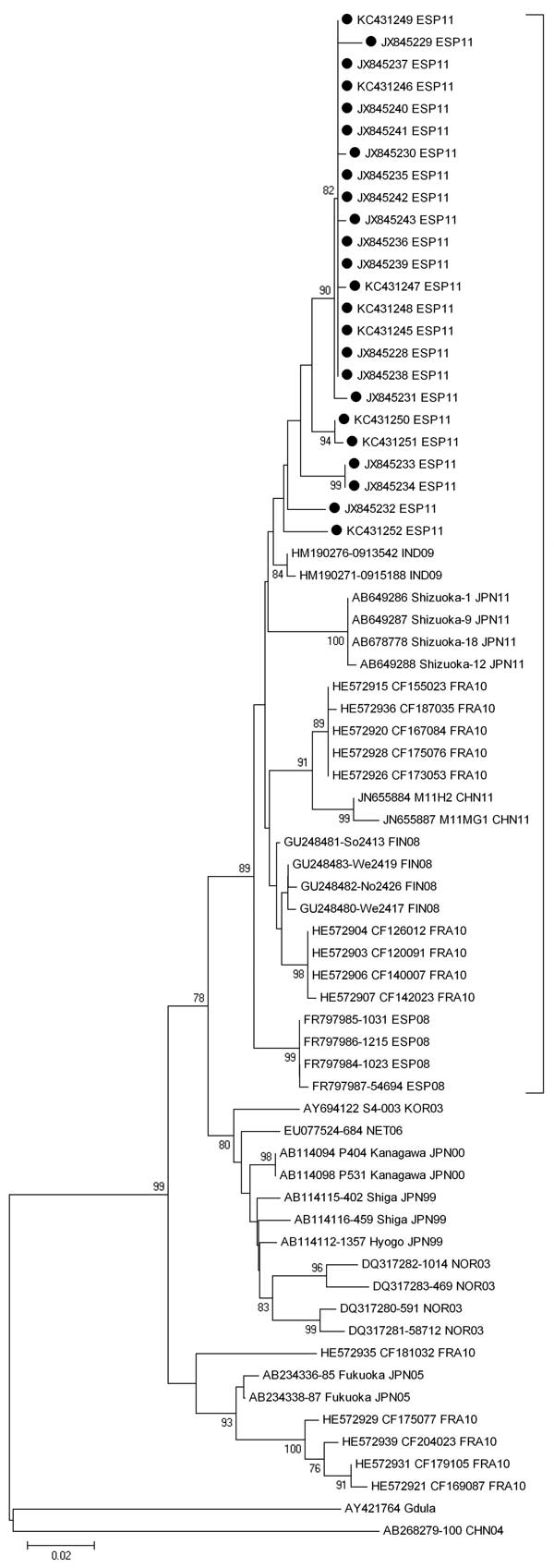
Figure. . . . Phylogenetic analysis of the partial viral protein 1 gene sequence (positions 2929–3348, based on strain Shizuoka-18, GenBank accession no. AB678778) of coxsackievirus A6 isolated from distinct patients with hand, foot, and mouth disease detected in Irun, Spain, April–September 2011, compared with the Gdula prototype strain and other representative strains. Black dots indicate the strains in this study (GenBank accession nos. JX845228–JX845243 and KC431245–431253). The tree was constructed by using the neighbor-joining method with 1,000 bootstrap replications and shows bootstrap values >75%. Genetic distances are based on pairwise analysis by using the Kimura 2-parameter method in MEGA5.1 software (www.megasoftware.net). Bracket indicates strains showing nucleotide identity >94% and detected in outbreaks during 2008–2011. Scale bar indicates the number of substitutions per nucleotide position.