Volume 19, Number 5—May 2013
Dispatch
Novel Lyssavirus in Bat, Spain
Figure
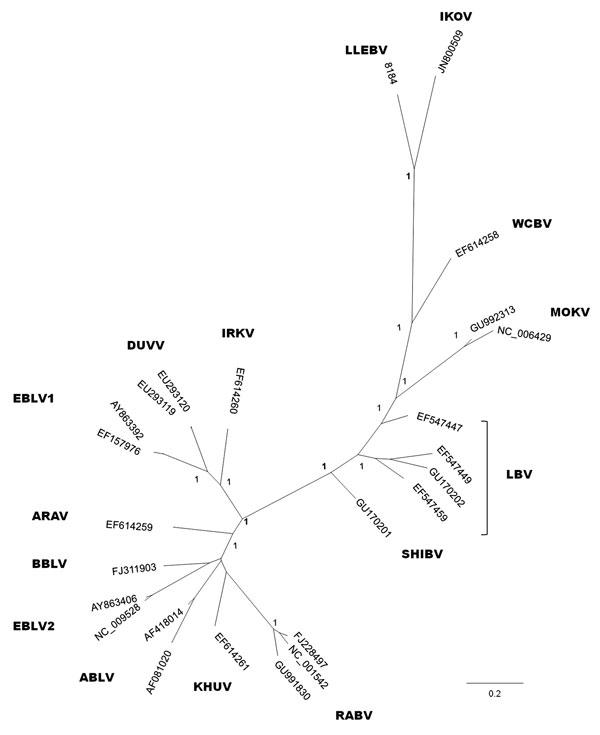
Figure. . Phylogenetic reconstruction based on the first 405 nt of the nucleoprotein gene, including all representative lyssaviruses. The tree was obtained by Bayesian inference, and the first 25% of trees were excluded from the analysis as burn-in. Node numbers indicate posterior probabilities. ARAV, Aravan virus; ABLV, Australian bat lyssavirus; BBLV, Bokeloh bat lyssavirus; DUVV, Duvenhage virus; EBLV-1 and EBLV-2, European bat lyssavirus types 1 and 2; IRKV, Irkut virus; KHUV, Khujand virus; LBV, Lagos bat virus (lineages A, B, C, and D); MOKV, Mokola virus; RABV, rabies virus; SHIBV, Shimoni bat virus; WCBV, West Caucasian bat virus; IKOV, Ikoma lyssavirus; LLEBV, Lleida bat lyssavirus (proposed). Scale bar indicates expected number of substitutions per site.
1These authors contributed equally to this article.