Volume 19, Number 8—August 2013
Dispatch
Recombinant Coxsackievirus A2 and Deaths of Children, Hong Kong, 2012
Figure 1
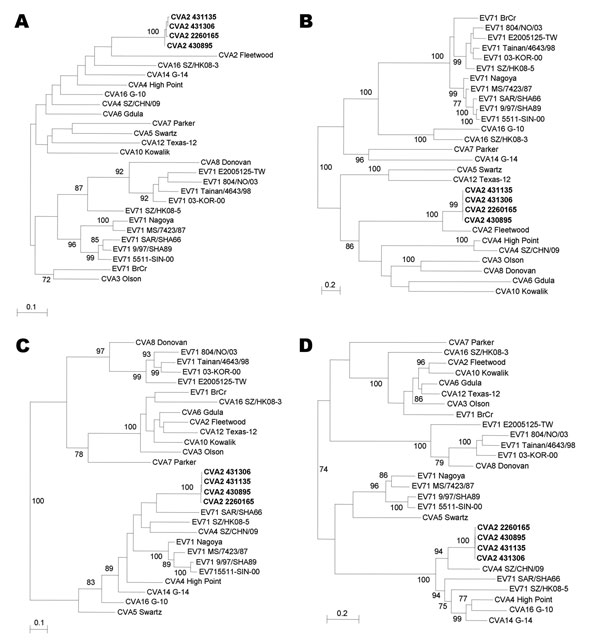
Figure 1. . Phylogenetic trees of A) 5′-untranslated region (UTR), B) capsid protein (P1), C) nonstructural protein 2 (P2), and D) nonstructural protein 3 (P3) regions of 4 coxsackievirus A2 (CVA2) strains, Hong Kong, 2012 and other human enterovirus (EV) A strains with complete genome sequences. Trees were inferred from data by using the maximum-likelihood method with bootstrap values calculated from 1,000 trees. Sequences for 758-nt positions in each 5′-UTR, 2,595 nt positions in each P1 region, 1,734 nt positions in each P2 region, and 2,259 nt positions in each P3 region were included in the analysis. Only bootstrap values >70% are shown. Scale bars indicate estimated number of nucleotide substitutions per 5 (B and D) or 10 (A and C) nucleotides. CVA2 strains isolated in this study are indicated in boldface. Virus strains (GenBank accession nos.) used were CVA2 Fleetwood (AY421760), CVA3 Olson (AY421761), CVA4 High Point (AY421762), CVA4 SZ/CHN/09 (HQ728260), CVA5 Swartz (AY421763), CVA6 Gdula (AY421764), CVA7 Parker (AY421765), CVA8 Donovan (AY421766), CVA10 Kowalik (AY421767), CVA12 Texas-12 (AY421768), CVA14 G-14 (AY421769), CVA16 G-10 (U05876), CVA16 SZ/HK08–3 (GQ279368), EV71 BrCr (U22521), EV71 Nagoya (AB482183), EV71 MS/7423/87 (U22522), EV71 SAR/SHA66 (AM396586), EV71 9/97/SHA89 (AJ586873), EV71 5511-SIN-00 (DQ341364), EV71 804/NO/03 (DQ452074), EV71 Tainan/4643/98 (AF304458), EV71 03-KOR-00 (DQ341356), EV71 SZ/HK08–5 (GQ279369), and EV71 E2005125-TW (EF063152).